For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGVSVIIRSLQEPVGRRRAVLRALCASRVPMSIAAIAGKLGVHPNTVRFHLDNLVADGQVERVEPGRGRP GRPPLMFRAVRRTDSTGTRRYRLLAEILASGLAAERDSRAMALSAGRAWGRQLEAPPAGADTEETIDHLV AVLDDLGFAPERRASNGRQQVGLRHCPFLELAETQAGVVCPVHLGIMRGALQTWAPVTVDRLDAFVEPDL CLAHFTPLEGAIR
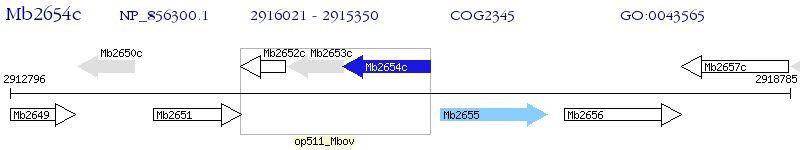
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2654c | - | - | 100% (223) | transcriptional regulatory protein |
| M. bovis AF2122 / 97 | Mb2651 | - | 1e-12 | 26.26% (198) | hypothetical protein Mb2651 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3487 | - | 4e-26 | 35.92% (206) | putative transcriptional regulator |
| M. tuberculosis H37Rv | Rv2621c | - | 1e-125 | 99.55% (224) | transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_00592 | - | 2e-06 | 28.27% (191) | putative DNA-binding protein |
| M. abscessus ATCC 19977 | MAB_0215 | - | 3e-14 | 32.32% (198) | transcriptional regulator |
| M. marinum M | MMAR_2081 | - | 1e-82 | 69.06% (223) | transcriptional regulator |
| M. avium 104 | MAV_3499 | - | 2e-77 | 68.78% (221) | hypothetical protein MAV_3499 |
| M. smegmatis MC2 155 | MSMEG_1125 | - | 3e-47 | 48.56% (208) | regulatory protein, ArsR |
| M. thermoresistible (build 8) | TH_1819 | - | 2e-45 | 45.71% (210) | regulatory protein, ArsR |
| M. ulcerans Agy99 | MUL_3263 | - | 7e-83 | 69.06% (223) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5848 | - | 1e-67 | 62.20% (209) | putative transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2654c|M.bovis_AF2122/97 -------------------MGVS------VIIRSLQEPVGRRRAVLRALC
Rv2621c|M.tuberculosis_H37Rv -------------------MGVS------VIIRSLQEPVGRRRAVLRALC
MMAR_2081|M.marinum_M -------------------MGE----GR--IARMGGEPAGRRRAVLRVLR
MUL_3263|M.ulcerans_Agy99 -------------------MGE----GR--IARMGGEPAGRRRAVLRVLR
MAV_3499|M.avium_104 -------------------MGDPLTKSRKLLAPSAGESAGRRRAVLRLLR
Mvan_5848|M.vanbaalenii_PYR-1 -----------------------------MSAASPGEPPGRRRDVLRTLK
MSMEG_1125|M.smegmatis_MC2_155 -----------------------------MDTDTSLPPRGRRDAILQVLR
TH_1819|M.thermoresistible__bu -----------------------------MPAVAEPDTGGRRNEILRLLQ
Mflv_3487|M.gilvum_PYR-GCK -----------------MNKEGLVLGPHPTRTVAGYPPLSRQRLCVLDYV
MAB_0215|M.abscessus_ATCC_1997 -------------------------------MTGSRGAGQQRARVLKVLQ
MLBr_00592|M.leprae_Br4923 MLNVHRGYPSLDRSPELRHTGVVKIRSELDAATTVAVSDGHTRRAVVRLL
. : :
Mb2654c|M.bovis_AF2122/97 ASRVPMSIAAIAGKLGVHPNTVRFHLDNLVADGQVERVEPG---RGRPGR
Rv2621c|M.tuberculosis_H37Rv ASRVPMSIAAIAGKLGVHPNTVRFHLDNLVADGQVERVEPG---RGRPGR
MMAR_2081|M.marinum_M ALGHPLSIVAIAEHLGVHPNTVRFHLDGLVDDGQVERVQPS---RRGPGR
MUL_3263|M.ulcerans_Agy99 ALGHPLSIVAIAEHLGVHPNTVRFHLDGLVDDGQVERVQPS---RRGPGR
MAV_3499|M.avium_104 ASPEPMSIAGIADVLGVHPNTVRFHLDSLVADGQVEHVELD---RKGPGR
Mvan_5848|M.vanbaalenii_PYR-1 AASGPMSIGAIADALDIHPNTVRFHLDRLVSQGQVERIEPD---RRRPGR
MSMEG_1125|M.smegmatis_MC2_155 ASPQPRAIAGIADELGVHPNTVRFHLEALARAGRVEQVVGE---AFGRGR
TH_1819|M.thermoresistible__bu ESDTARGVAGLADELRVHPNTVRFHLNALVRTGDVEQVSGG---TAGPGR
Mflv_3487|M.gilvum_PYR-GCK RTHAPVRITAVAAALDLHPNTVREHLDALTGHGLAERVTET---PSKRGR
MAB_0215|M.abscessus_ATCC_1997 EAAAPVGAREVADSLGLHITTVRFHLKTLEEQGHIVRRGGTG--EQRAGR
MLBr_00592|M.leprae_Br4923 LESGSITAGEISDRLGLSAAGVRRHLDVLIEMGDAESATAAPWQQVGRGR
. :: * : ** **. * * **
Mb2654c|M.bovis_AF2122/97 PPLMFRAVRRTDSTGTRRYRLLAEILASGLAAER-DSRAMALSAGRAWGR
Rv2621c|M.tuberculosis_H37Rv PPLMFRAVRRTDSTGTRRYRLLAEILASGLAAER-DSRAMALSAGRAWGR
MMAR_2081|M.marinum_M PPLMFRAVQQMDRGGARHYQLLAEILSNGLAAES-DSVDKALAAGRAWGE
MUL_3263|M.ulcerans_Agy99 PPLMFRAVQQMDRGGARHYQLLAEILSNGLAAES-DSVDKALAAGRAWGE
MAV_3499|M.avium_104 PPLMFRAVRQMDRGGTRHYRLLAEILAKAFAAET-DPAPKALAAGRAWGQ
Mvan_5848|M.vanbaalenii_PYR-1 PPQMFRAMPQMDRAGPRHYRLLAEMLAMSLAADG-DPPTRAQAGGRAWGR
MSMEG_1125|M.smegmatis_MC2_155 PPVLYRASRRMDPSGPTNYRLLASILTGYVARHP-DAAAVAADLGRSTSP
TH_1819|M.thermoresistible__bu PPVLFRAVRRMDPAGPTNYRLLADIMTSHFETNTDDPRAAAAELGRRWGP
Mflv_3487|M.gilvum_PYR-GCK PAALYRPSAADPAIPAQDYAGLATALAGHIARTSTQPERDARAAGASWGH
MAB_0215|M.abscessus_ATCC_1997 PSLSYAVAPRLD------YADVVALFAVHLGGTAAERESRAALVGADLAH
MLBr_00592|M.leprae_Br4923 PAKHYRLTAVGRAKLGHAYDTLASAAMRQLREIGGEDAVRTFARQRIDAI
*. : * :. . : : .
Mb2654c|M.bovis_AF2122/97 QLEA-----PPAGADTEETIDHLVAVLDDLGFAP-ERRAS--NGRQQVGL
Rv2621c|M.tuberculosis_H37Rv QLEA-----PPAGADTEETIDHLVAVLDDLGFAP-ERRAS--NGRQQVGL
MMAR_2081|M.marinum_M RLQA-----PTTAHSAEKSIDHLVGVLDGLGFAP-ERRVS--DGQQQVAL
MUL_3263|M.ulcerans_Agy99 RLQA-----PTTAHSAEKSIDHLVGVLDGLGFAP-ERRVS--DGQQQVAL
MAV_3499|M.avium_104 QLDAERRRLPRDAAGAEQAIAQLVDVLDELGFAP-ERRVI--DGEQQVGL
Mvan_5848|M.vanbaalenii_PYR-1 ELNVRAP--RRKQSGAEEAVDRLVEVLDDVGFVP-ERLGA--GEKQQIGL
MSMEG_1125|M.smegmatis_MC2_155 GLVTTA---PHGPSTRTQAVTELVDLLAELGFEP-EPADG--RRAREIRL
TH_1819|M.thermoresistible__bu SLVRRQ---PQGEPSRQETLAELTDVLADVGFRP-DPPVG--RSRPRITL
Mflv_3487|M.gilvum_PYR-GCK ELCD-------DTVTTDDPRAAVLTVLARLGFGP-DADGP--DGS--IAL
MAB_0215|M.abscessus_ATCC_1997 RVNVAR------RRTPLQVTDLVVETLGELGFTVRSTLMS--FGRVTVQI
MLBr_00592|M.leprae_Br4923 LADVEP---AGGGADVEAAANRVASALSDAGYVATTARVGGPIHGVEICQ
: * *: :
Mb2654c|M.bovis_AF2122/97 RHCPFLELAETQAGVVCPVHLGIMRGALQTW-APVTVD-RLDAFVEP--D
Rv2621c|M.tuberculosis_H37Rv RHCPFLELAETQAGVVCPVHLGIMRGALQTWGAPVTVD-RLDAFVEP--D
MMAR_2081|M.marinum_M RHCPFLELAENRSGVVCPIHLGLMRGALESWGAPVTVE-RLDPFVEP--D
MUL_3263|M.ulcerans_Agy99 RHCPFLELAENRSGVVCPIHLGLMRGALESWGAPVTVE-RLDPFVEP--D
MAV_3499|M.avium_104 RHCPFLELAENSSNVVCPVHLGLMQGAMESWGAPVSVD-RLDPFVEP--D
Mvan_5848|M.vanbaalenii_PYR-1 RHCPFLELAETQSRVVCGIHLGLMQGVLERQGAPVTVD-RLEPFAQP--D
MSMEG_1125|M.smegmatis_MC2_155 RHCPFQNLAEEHGEVTCALHLGLMQGALRDLQAPVTVD-RLEPFVEP--D
TH_1819|M.thermoresistible__bu RHCPFYELVTRYGETICGLHLGLMQGVLAAIRGPVSVA-GLDPLVEP--D
Mflv_3487|M.gilvum_PYR-GCK RSCPLLEAARRYPAIVCQVHLGIVEGVLERFGAVTDTAPELVAFAEP--G
MAB_0215|M.abscessus_ATCC_1997 CSCPLAEIATTAPEVVRGIQRGLIQEVLDVNADAIGGQFQVTVSPDAGHG
MLBr_00592|M.leprae_Br4923 HHCPVSHVAEEFP--------ELCETEQQAIAEVLGTHVQRLAMIVNGDC
**. . . : .
Mb2654c|M.bovis_AF2122/97 LCLAHFTPLEGAIR-----
Rv2621c|M.tuberculosis_H37Rv LCLAHFTPLEGAIR-----
MMAR_2081|M.marinum_M LCLAHLS-PEGALS-----
MUL_3263|M.ulcerans_Agy99 LCLAHLS-PEGALS-----
MAV_3499|M.avium_104 LCLAHLT-LQGADR-----
Mvan_5848|M.vanbaalenii_PYR-1 LCLAHLT-LRGRPPQA---
MSMEG_1125|M.smegmatis_MC2_155 LCVAHLAPR----------
TH_1819|M.thermoresistible__bu LCVAHLSTTPGWRRE----
Mflv_3487|M.gilvum_PYR-GCK ACRLFLPDPVVPERNRP--
MAB_0215|M.abscessus_ATCC_1997 DCTVNLALVPLGKESWPWT
MLBr_00592|M.leprae_Br4923 ACTTHVPLMSVPSKT----
* ..