For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GEGRIARMGGEPAGRRRAVLRVLRALGHPLSIVAIAEHLGVHPNTVRFHLDGLVDDGQVERVQPSRRGPG RPPLMFRAVQQMDRGGARHYQLLAEILSNGLAAESDSVDKALAAGRAWGERLQAPTTAHSAEKSIDHLVG VLDGLGFAPERRVSDGQQQVALRHCPFLELAENRSGVVCPIHLGLMRGALESWGAPVTVERLDPFVEPDL CLAHLSPEGALS*
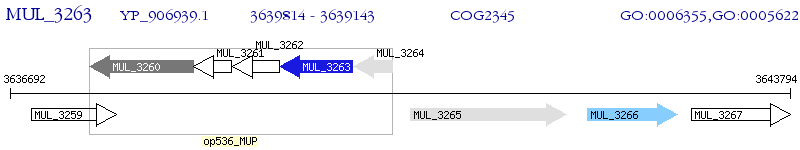
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3263 | - | - | 100% (223) | transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2654c | - | 7e-83 | 69.06% (223) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_3487 | - | 4e-25 | 36.10% (205) | putative transcriptional regulator |
| M. tuberculosis H37Rv | Rv2621c | - | 6e-85 | 69.51% (223) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0215 | - | 3e-11 | 30.65% (186) | transcriptional regulator |
| M. marinum M | MMAR_2081 | - | 1e-127 | 100.00% (223) | transcriptional regulator |
| M. avium 104 | MAV_3499 | - | 2e-87 | 71.49% (221) | hypothetical protein MAV_3499 |
| M. smegmatis MC2 155 | MSMEG_1125 | - | 2e-49 | 48.56% (208) | regulatory protein, ArsR |
| M. thermoresistible (build 8) | TH_1819 | - | 2e-45 | 43.54% (209) | regulatory protein, ArsR |
| M. vanbaalenii PYR-1 | Mvan_5848 | - | 1e-71 | 62.91% (213) | putative transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2654c|M.bovis_AF2122/97 --MGVS------VIIRSLQEPVGRRRAVLRALCASRVPMSIAAIAGKLGV
Rv2621c|M.tuberculosis_H37Rv --MGVS------VIIRSLQEPVGRRRAVLRALCASRVPMSIAAIAGKLGV
MUL_3263|M.ulcerans_Agy99 --MGE----GR--IARMGGEPAGRRRAVLRVLRALGHPLSIVAIAEHLGV
MMAR_2081|M.marinum_M --MGE----GR--IARMGGEPAGRRRAVLRVLRALGHPLSIVAIAEHLGV
MAV_3499|M.avium_104 --MGDPLTKSRKLLAPSAGESAGRRRAVLRLLRASPEPMSIAGIADVLGV
Mvan_5848|M.vanbaalenii_PYR-1 ------------MSAASPGEPPGRRRDVLRTLKAASGPMSIGAIADALDI
MSMEG_1125|M.smegmatis_MC2_155 ------------MDTDTSLPPRGRRDAILQVLRASPQPRAIAGIADELGV
TH_1819|M.thermoresistible__bu ------------MPAVAEPDTGGRRNEILRLLQESDTARGVAGLADELRV
Mflv_3487|M.gilvum_PYR-GCK MNKEGLVLGPHPTRTVAGYPPLSRQRLCVLDYVRTHAPVRITAVAAALDL
MAB_0215|M.abscessus_ATCC_1997 --------------MTGSRGAGQQRARVLKVLQEAAAPVGAREVADSLGL
. :: : . :* * :
Mb2654c|M.bovis_AF2122/97 HPNTVRFHLDNLVADGQVERVEPG-RGRPGRPPLMFRAVRRTDSTGTRRY
Rv2621c|M.tuberculosis_H37Rv HPNTVRFHLDNLVADGQVERVEPG-RGRPGRPPLMFRAVRRTDSTGTRRY
MUL_3263|M.ulcerans_Agy99 HPNTVRFHLDGLVDDGQVERVQPS-RRGPGRPPLMFRAVQQMDRGGARHY
MMAR_2081|M.marinum_M HPNTVRFHLDGLVDDGQVERVQPS-RRGPGRPPLMFRAVQQMDRGGARHY
MAV_3499|M.avium_104 HPNTVRFHLDSLVADGQVEHVELD-RKGPGRPPLMFRAVRQMDRGGTRHY
Mvan_5848|M.vanbaalenii_PYR-1 HPNTVRFHLDRLVSQGQVERIEPD-RRRPGRPPQMFRAMPQMDRAGPRHY
MSMEG_1125|M.smegmatis_MC2_155 HPNTVRFHLEALARAGRVEQVVGE-AFGRGRPPVLYRASRRMDPSGPTNY
TH_1819|M.thermoresistible__bu HPNTVRFHLNALVRTGDVEQVSGG-TAGPGRPPVLFRAVRRMDPAGPTNY
Mflv_3487|M.gilvum_PYR-GCK HPNTVREHLDALTGHGLAERVTET-PSKRGRPAALYRPSAADPAIPAQDY
MAB_0215|M.abscessus_ATCC_1997 HITTVRFHLKTLEEQGHIVRRGGTGEQRAGRPSLSYAVAPRLD------Y
* .*** **. * * : ***. : *
Mb2654c|M.bovis_AF2122/97 RLLAEILASGLAAER-DSRAMALSAGRAWGRQLEA-----PPAGADTEET
Rv2621c|M.tuberculosis_H37Rv RLLAEILASGLAAER-DSRAMALSAGRAWGRQLEA-----PPAGADTEET
MUL_3263|M.ulcerans_Agy99 QLLAEILSNGLAAES-DSVDKALAAGRAWGERLQA-----PTTAHSAEKS
MMAR_2081|M.marinum_M QLLAEILSNGLAAES-DSVDKALAAGRAWGERLQA-----PTTAHSAEKS
MAV_3499|M.avium_104 RLLAEILAKAFAAET-DPAPKALAAGRAWGQQLDAERRRLPRDAAGAEQA
Mvan_5848|M.vanbaalenii_PYR-1 RLLAEMLAMSLAADG-DPPTRAQAGGRAWGRELNVRAP--RRKQSGAEEA
MSMEG_1125|M.smegmatis_MC2_155 RLLASILTGYVARHP-DAAAVAADLGRSTSPGLVTTA---PHGPSTRTQA
TH_1819|M.thermoresistible__bu RLLADIMTSHFETNTDDPRAAAAELGRRWGPSLVRRQ---PQGEPSRQET
Mflv_3487|M.gilvum_PYR-GCK AGLATALAGHIARTSTQPERDARAAGASWGHELCD-------DTVTTDDP
MAB_0215|M.abscessus_ATCC_1997 ADVVALFAVHLGGTAAERESRAALVGADLAHRVNVAR------RRTPLQV
:. :: . : * * . : .
Mb2654c|M.bovis_AF2122/97 IDHLVAVLDDLGFAP-ERRASNGRQQVGLRHCPFLELAETQAGVVCPVHL
Rv2621c|M.tuberculosis_H37Rv IDHLVAVLDDLGFAP-ERRASNGRQQVGLRHCPFLELAETQAGVVCPVHL
MUL_3263|M.ulcerans_Agy99 IDHLVGVLDGLGFAP-ERRVSDGQQQVALRHCPFLELAENRSGVVCPIHL
MMAR_2081|M.marinum_M IDHLVGVLDGLGFAP-ERRVSDGQQQVALRHCPFLELAENRSGVVCPIHL
MAV_3499|M.avium_104 IAQLVDVLDELGFAP-ERRVIDGEQQVGLRHCPFLELAENSSNVVCPVHL
Mvan_5848|M.vanbaalenii_PYR-1 VDRLVEVLDDVGFVP-ERLGAGEKQQIGLRHCPFLELAETQSRVVCGIHL
MSMEG_1125|M.smegmatis_MC2_155 VTELVDLLAELGFEP-EPADGRRAREIRLRHCPFQNLAEEHGEVTCALHL
TH_1819|M.thermoresistible__bu LAELTDVLADVGFRP-DPPVGRSRPRITLRHCPFYELVTRYGETICGLHL
Mflv_3487|M.gilvum_PYR-GCK RAAVLTVLARLGFGP-DADGPDGS--IALRSCPLLEAARRYPAIVCQVHL
MAB_0215|M.abscessus_ATCC_1997 TDLVVETLGELGFTVRSTLMSFGRVTVQICSCPLAEIATTAPEVVRGIQR
: * :** . : : **: : . ::
Mb2654c|M.bovis_AF2122/97 GIMRGALQTW-APVTVD-RLDAFVEP--DLCLAHFTPLEGAIR-----
Rv2621c|M.tuberculosis_H37Rv GIMRGALQTWGAPVTVD-RLDAFVEP--DLCLAHFTPLEGAIR-----
MUL_3263|M.ulcerans_Agy99 GLMRGALESWGAPVTVE-RLDPFVEP--DLCLAHLS-PEGALS-----
MMAR_2081|M.marinum_M GLMRGALESWGAPVTVE-RLDPFVEP--DLCLAHLS-PEGALS-----
MAV_3499|M.avium_104 GLMQGAMESWGAPVSVD-RLDPFVEP--DLCLAHLT-LQGADR-----
Mvan_5848|M.vanbaalenii_PYR-1 GLMQGVLERQGAPVTVD-RLEPFAQP--DLCLAHLT-LRGRPPQA---
MSMEG_1125|M.smegmatis_MC2_155 GLMQGALRDLQAPVTVD-RLEPFVEP--DLCVAHLAPR----------
TH_1819|M.thermoresistible__bu GLMQGVLAAIRGPVSVA-GLDPLVEP--DLCVAHLSTTPGWRRE----
Mflv_3487|M.gilvum_PYR-GCK GIVEGVLERFGAVTDTAPELVAFAEP--GACRLFLPDPVVPERNRP--
MAB_0215|M.abscessus_ATCC_1997 GLIQEVLDVNADAIGGQFQVTVSPDAGHGDCTVNLALVPLGKESWPWT
*::. .: : :. . * :.