For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FDRFRHTGQPTSKRKRPSLPPTLYGTLPVSPTIALGTALFPVAVRAIGALLILKKPPENHVDRTLKLVVK DRWIEAKDQDVASLVLASPDGDRLPPWHPGAHLDLLLPSGRMRQYSLCGDVSDPYHYRIAVRRIPGGGGG SIEVHDALQIGATVSILGPRNAFPLAMTQPGPRKLHFVAGGIGITPILSMIAFAEQLGKPWTMVYTGRHR DSLPFLNELKRFGDRVIIRTDDQSGLPTADDLLPGVGENDAVYCCGPAPMLAVLQRRLLEMPSVELHFER FAAAPVVDGHPFEVQLGPGGPVLDVPADRTALDVIKKRLPHVAYSCQQGFCGTCKVNVLAGTVDHRDQIL TESQRDEGQILTCVSRGEGRLVLDLPAE*
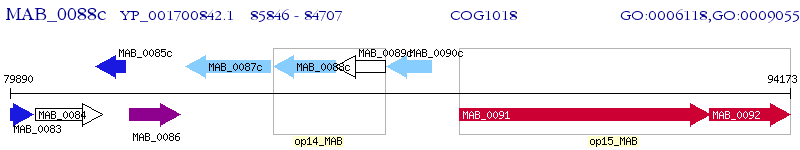
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0088c | - | - | 100% (379) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_4020c | - | e-112 | 58.12% (351) | oxidoreductase |
| M. abscessus ATCC 19977 | MAB_2381 | - | 1e-52 | 39.44% (322) | oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2798c | - | 3e-76 | 49.84% (317) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_3426 | - | 9e-95 | 53.26% (353) | ferredoxin |
| M. tuberculosis H37Rv | Rv2776c | - | 3e-76 | 49.84% (317) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1933 | - | 1e-79 | 47.79% (362) | oxidoreductase |
| M. avium 104 | MAV_3669 | - | 3e-77 | 51.72% (319) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_3676 | - | 2e-97 | 51.77% (367) | phenoxybenzoate dioxygenase beta subunit |
| M. thermoresistible (build 8) | TH_3804 | - | 1e-101 | 56.84% (329) | PUTATIVE ferredoxin |
| M. ulcerans Agy99 | MUL_2159 | - | 3e-78 | 47.51% (362) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_3180 | - | 6e-97 | 52.11% (380) | ferredoxin |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3676|M.smegmatis_MC2_155 ---------MLSQYRQL------------PPSTNGRFRHDPMLAMAQIAI
TH_3804|M.thermoresistible__bu -------MRMLARLRAT------------PPSSSGRIRHDLLLGVADATI
Mflv_3426|M.gilvum_PYR-GCK ------------------------------MGVLGRLRYDVTIGLADAAL
Mvan_3180|M.vanbaalenii_PYR-1 ------MTRLFSQFRQT------------PPSPSGRLRHDVMLGIAETAI
MAB_0088c|M.abscessus_ATCC_199 ---------MFDRFRHTGQPTSKRKRPSLPPTLYGTLPVSPTIALGTALF
Mb2798c|M.bovis_AF2122/97 --------------------------------------------------
Rv2776c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1933|M.marinum_M ------MAVSESSWTSR------------PADLYGRSHRDR-FATALWGV
MUL_2159|M.ulcerans_Agy99 ------MAVSESSWTSR------------PADLYGRSHRDR-FATALWGV
MAV_3669|M.avium_104 MSIRRAASRAPSIWASR------------PADLYGRRDHDR-FTAVLWGV
MSMEG_3676|M.smegmatis_MC2_155 KTMWGVTRHIRRPSPPPA--LDRTIGLRVVDRRVVAHDQDVIALTLAADD
TH_3804|M.thermoresistible__bu SSLWKVSEVLRTPRLPAV--DAEPITLTVRDRRVVAHDQDVIALTLVAAD
Mflv_3426|M.gilvum_PYR-GCK HGMFATSAAMRRIRPPDG--VPETFPLTVTSRRVVARDQDVVELTLSGDD
Mvan_3180|M.vanbaalenii_PYR-1 AGMFAASAAVRRVRMPAA--GPDTIPLTVTGRRVVAHDQDVVELTLAGDR
MAB_0088c|M.abscessus_ATCC_199 PVAVRAIGALLILKKPPENHVDRTLKLVVKDRWIEAKDQDVASLVLASPD
Mb2798c|M.bovis_AF2122/97 --------------------MRRTNPAVVTKRELVAP--DVVALTLADPG
Rv2776c|M.tuberculosis_H37Rv --------------------MRRTNPAVVTKRELVAP--DVVALTLADPG
MMAR_1933|M.marinum_M RTMFGGFARFSRWNPSRVTPVGRTHRARVVERELVAP--DVVALTLADPD
MUL_2159|M.ulcerans_Agy99 RTMFGGFARFSRWNPSRVTPVGRTHRARVVERELVAP--DVVALTLADPD
MAV_3669|M.avium_104 RAVFEGFASVSRWHPARVKPVQRKLTAVVTKREFAAP--DVVALTLADPD
* * . * ** *.*
MSMEG_3676|M.smegmatis_MC2_155 GGALPRWYPGAHIDLHLPSGRIRQYSLCGDPAAG-TYRIAVRRIPEGGGG
TH_3804|M.thermoresistible__bu GGQLPPWRPGAHIDVTLPSGRVRQYSLCGDPAVRDRYRIAVRRIPDGGGG
Mflv_3426|M.gilvum_PYR-GCK ---LPRWHPGAHLDLHLPSGRIRQYSLCGEPTAG-HYRVAVRRIPDGGGG
Mvan_3180|M.vanbaalenii_PYR-1 ---LPRWFPGAHLDVHLPSGRVRQYSLCGDPDSA-EYRIAVRRIPDGGGG
MAB_0088c|M.abscessus_ATCC_199 GDRLPPWHPGAHLDLLLPSGRMRQYSLCGDVSDPYHYRIAVRRIPGGGGG
Mb2798c|M.bovis_AF2122/97 GGLLPAWSPGGHIDVQLPSGRRRQYSLCGVPGRRTDYRIAIRRIADGGGG
Rv2776c|M.tuberculosis_H37Rv GGLLPAWSPGGHIDVQLPSGRRRQYSLCGVPGRRTDYRIAIRRIADGGGG
MMAR_1933|M.marinum_M GGLLPSWSPGGHIDVQLPSGRRRQYSLCGPPGRRTDYRIAVRRIADGGGG
MUL_2159|M.ulcerans_Agy99 GGLLPSWSPGGHIDVQLPSRRRRQYSLCGPPGRRTDYRIAVRRIADGGGG
MAV_3669|M.avium_104 GGLLPSWTPGAHIDVLLPSGRRRQYSLCGPPGHRGEYRIAVRRIADGGGG
** * **.*:*: *** * ******* **:*:***. ****
MSMEG_3676|M.smegmatis_MC2_155 SVEIHDTVHIGSRLTTHGPRNAFPLTVPGYGSPTQRFRFIAGGIGITPIL
TH_3804|M.thermoresistible__bu SVEVHDALHLGARVTTHGPRNAFPLSVPGYGSPARRLRFIAGGIGITPIL
Mflv_3426|M.gilvum_PYR-GCK SVEVHDALPVGSTVRTHGPRNAFPLAVPGYGSPARRFRFIAGGIGITPIL
Mvan_3180|M.vanbaalenii_PYR-1 SVEVHDALEIGSTVHTHGPRNAFPLTVPGYGSPARRFRFIAGGIGITPIL
MAB_0088c|M.abscessus_ATCC_199 SIEVHDALQIGATVSILGPRNAFPLAMTQPGP--RKLHFVAGGIGITPIL
Mb2798c|M.bovis_AF2122/97 SIEMHEAFDVGDTCEFEGPRNAFHL-----GLAERDVLFVIGGIGVTPIL
Rv2776c|M.tuberculosis_H37Rv SIEMHEAFDVGDTCEFEGPRNAFHL-----GLAERDVLFVIGGIGVTPIL
MMAR_1933|M.marinum_M SIEMHAAYQPGDILVFEGPRNAFYL-----GAAEHDMLFVIGGIGVTPIL
MUL_2159|M.ulcerans_Agy99 SIEMHAAYQPGDILVFEGPRNAFYL-----GAAEHDMLFVIGGIGVTPIL
MAV_3669|M.avium_104 SIEMHEAFGVGDTVEFEGPRNAFYL-----GTAEHDVLFVIGGIGVTPIL
*:*:* : * ****** * * : . *: ****:****
MSMEG_3676|M.smegmatis_MC2_155 PMLDLAQRLGVDWSMIYAGRHPDSLPFLSELQRYG-DRIGIRTDDRDG-L
TH_3804|M.thermoresistible__bu PMLAEAQAFGVPWSMIYAGRSRDSLPFLEELTRYG-DRVRVRTDDVAG-L
Mflv_3426|M.gilvum_PYR-GCK PMLALARSLGVDWSMVYTGRSADALPFVDEVAAFG-SPVEIRTDDRRG-L
Mvan_3180|M.vanbaalenii_PYR-1 PMLGLARRLGVDWSMVYTGRSVDSLPFVDEVTAFG-AAVDIRTDDRHG-L
MAB_0088c|M.abscessus_ATCC_199 SMIAFAEQLGKPWTMVYTGRHRDSLPFLNELKRFG-DRVIIRTDDQSG-L
Mb2798c|M.bovis_AF2122/97 PMIRAAEQRGIDWRAIYAGRGREYMPFLDEVVAVAPGRVTVWADDEHGRF
Rv2776c|M.tuberculosis_H37Rv PMIRAAEQRGIDWRAIYAGRGREYMPFLDEVVAVAPGRVTVWADDEHGRF
MMAR_1933|M.marinum_M PMVALAHQRGVNWRAMYAGRSREYMPLLDELVAMAPERVSVWADDEHGRF
MUL_2159|M.ulcerans_Agy99 PMVALAHQRGVNWRAVYAGRSREYMPLLDELVAMAPEGVSVWADDEHGRF
MAV_3669|M.avium_104 PMIRVAAGRGIDWRAIYTGRSREYMPLLDEVVSVDPQRVTVWADDEHGRF
.*: * * * :*:** : :*::.*: : : :** * :
MSMEG_3676|M.smegmatis_MC2_155 PGASDLLGECPGGTTVYACGPAPMLTMLRTALVGRDDVELHFERFAAPPV
TH_3804|M.thermoresistible__bu PTAAELLGACPDGTAVYACGPPAMLTAIRAELAGRRNVELHFERFAAPPV
Mflv_3426|M.gilvum_PYR-GCK PTAAELLGDCPADTSVYACGPAPMLTAVRDALVGRDDVELHFERFAAPPV
Mvan_3180|M.vanbaalenii_PYR-1 PAAAELLGDCADGTAVYACGPAPMLTAVRAALVGRDDVELHFERFAAPPV
MAB_0088c|M.abscessus_ATCC_199 PTADDLLPGVGENDAVYCCGPAPMLAVLQRRLLEMPSVELHFERFAAAPV
Mb2798c|M.bovis_AF2122/97 ASVDELLAGAGPTTAVYVCGPPGMLEAVRVARNQHADAPLHYERFSPPPV
Rv2776c|M.tuberculosis_H37Rv ASVDELLAGAGPTTAVYVCGPPGMLEAVRVARNQHADAPLHYERFSPPPV
MMAR_1933|M.marinum_M AQVEDLVAGAGPKTAVYVCGPPPMLEAVRAARAEYPCAPLHYERFSPPPV
MUL_2159|M.ulcerans_Agy99 AQVEDLVAGAGLKTAVYVCGPPPMLEAVRAARAEYPCAPLHYERFSPPPV
MAV_3669|M.avium_104 PTAAELLAAAGPATAVYVCGPSAMLEAVWIARNEHAGAPLHYERFSPAPV
. . :*: :** ***. ** : . **:***:..**
MSMEG_3676|M.smegmatis_MC2_155 VDGTEFSVRIASTGQTVPVAADETLLAALNRAGVSAPYSCQQGFCGTCRV
TH_3804|M.thermoresistible__bu VDGRPFSVRIASTGAEVPVGADETLLAALGRAGVQAPYSCQQGFCGTCRT
Mflv_3426|M.gilvum_PYR-GCK VDGRPFR--VAAAGVTVDVGVDETLLAALGRAGVTTPYSCRQGFCGTCRT
Mvan_3180|M.vanbaalenii_PYR-1 TDGRPFQATVASTGQQVDVGADETLLAALRRAGVDASYSCQQGFCGTCRT
MAB_0088c|M.abscessus_ATCC_199 VDGHPFEVQLGPGGPVLDVPADRTALDVIKKRLPHVAYSCQQGFCGTCKV
Mb2798c|M.bovis_AF2122/97 VDGVPFELELARSRRVLRVPANRSALDVMLDWDPTTAYSCQQGFCGTCKV
Rv2776c|M.tuberculosis_H37Rv VDGVPFELELARSRRVLRVPANRSALDVMLDWDPTTAYSCQQGFCGTCKV
MMAR_1933|M.marinum_M VDGVPFELELARTGEVLSIPANRTALDVMLDRDPTTAYSCQQGFCGTCKV
MUL_2159|M.ulcerans_Agy99 VDGVPFELELARTGEVLSIPANRTALDVMLDRDPTTAYSCQQGFCGTCKV
MAV_3669|M.avium_104 VDGVPFELELARSRQVLAVPANRTALDVMLDRDATTPYSCRQGFCGTCRV
.** * :. : : .:.: * .: ..***:*******:.
MSMEG_3676|M.smegmatis_MC2_155 RALPRDGDGAVQHRDTLLTEPERAAGYLLTCVSRAGNGQRLTIEL---
TH_3804|M.thermoresistible__bu RVL----DGTVEHRDRLLTEPERAAGLMLTCVSRAPAGERLTLDL---
Mflv_3426|M.gilvum_PYR-GCK RVL----SGEVDHRDTLLTDAERAAGLMLPCVSRAAEGTVLALDL---
Mvan_3180|M.vanbaalenii_PYR-1 RVL----AGSVEHRDTLLTGPERAAGLMLTCVSRAGDGSGLTLDL---
MAB_0088c|M.abscessus_ATCC_199 NVL----AGTVDHRDQILTESQRDEGQILTCVSRG-EG-RLVLDLPAE
Mb2798c|M.bovis_AF2122/97 RVL----AGQVDRRGRIIEGDNE----MLVCVSRAVSG-RVVIDA---
Rv2776c|M.tuberculosis_H37Rv RVL----AGQVDRRGRIIEGDNE----MLVCVSRAVSG-RVVIDA---
MMAR_1933|M.marinum_M KVL----AGQVDHRGRAAQGDDE----MLVCVSRGHSA-RVVIDA---
MUL_2159|M.ulcerans_Agy99 KVL----AGQVDHRGRAAQGDDE----MLVCVSRGHSA-RVVIDA---
MAV_3669|M.avium_104 KVL----AGQVDHRGRTDPGQDG----MLVCVSRADGG-RLVIDA---
..* * *::*. : :* ****. . :.::