For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSPAEREFDIVLYGATGFSGKLTAEHLAHSGSTARIALAGRSSERLRGVRMMLGPNAADWPLILADASQP LTLEAMAARAQVVLTTVGPYTRYGLPLVAACAKAGTDYADLTGELMFCRNSIDLYHKQAADTGARIILAC GFDSIPSDLNVYQLYRRSVEDGTGELCDTDLVLRSFSQRWVSGGSVATYSEAMRTASSDPEARRLVTDPY TLTTDRGAEPELGAQPDFLRRPGRDLAPELAGFWTGGFVQAPFNTRIVRRSNALQEWAYGRRFRYSETMS LGKSMAAPILAAAVTGTVAGTIGLGNKYFDRLPRRLVERVTPKPGTGPSRKTQERGHYTFETYTTTTTGA RYRATFAHNVDAYKSTAVLLAQSGLALALDRDRLAELRGVLTPAAAMGDALLARLPGAGVVMGTTRLS
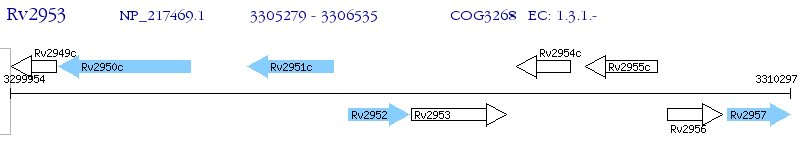
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2953 | - | - | 100% (418) | hypothetical protein Rv2953 |
| M. tuberculosis H37Rv | Rv2449c | - | e-158 | 68.12% (414) | hypothetical protein Rv2449c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2977 | - | 0.0 | 100.00% (418) | hypothetical protein Mb2977 |
| M. gilvum PYR-GCK | Mflv_2628 | - | 1e-156 | 66.11% (419) | saccharopine dehydrogenase |
| M. leprae Br4923 | MLBr_00129 | - | 0.0 | 86.60% (418) | hypothetical protein MLBr_00129 |
| M. abscessus ATCC 19977 | MAB_4339c | - | 1e-138 | 62.98% (416) | hypothetical protein MAB_4339c |
| M. marinum M | MMAR_1757 | - | 0.0 | 83.21% (417) | hypothetical protein MMAR_1757 |
| M. avium 104 | MAV_1723 | - | 1e-159 | 68.84% (414) | saccharopine dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4632 | - | 1e-156 | 66.91% (414) | saccharopine dehydrogenase |
| M. thermoresistible (build 8) | TH_0608 | - | 1e-161 | 67.78% (419) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2000 | - | 0.0 | 82.73% (417) | hypothetical protein MUL_2000 |
| M. vanbaalenii PYR-1 | Mvan_3953 | - | 1e-152 | 65.54% (415) | saccharopine dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2628|M.gilvum_PYR-GCK MDMSE-NDREHDIVLYGATGFVGKLTAQYLAS--AGTGARIALAGRSTDR
Mvan_3953|M.vanbaalenii_PYR-1 MSANEAHDREHDIVVYGATGFVGKLTAQYLAA--AGAGARIALAGRSTDR
TH_0608|M.thermoresistible__bu MTAAQ---REFDIVLYGATGFVGKLTARYLAS--AGVGARIALAGRSEQK
MSMEG_4632|M.smegmatis_MC2_155 MTQ-----REFDLVLYGATGFAGKLTAQYLAG--AAGDARIALAGRSAEK
MAV_1723|M.avium_104 MTATP---REFDIVLYGATGFVGKLTAEYLAG--AAPDKRIALAGRSTEK
MAB_4339c|M.abscessus_ATCC_199 MSR------EHDIVLYGATGYVGKLTALYLAGRVEATGARIALAGRSTEK
Rv2953|M.tuberculosis_H37Rv MSPAE---REFDIVLYGATGFSGKLTAEHLAH--SGSTARIALAGRSSER
Mb2977|M.bovis_AF2122/97 MSPAE---REFDIVLYGATGFSGKLTAEHLAH--SGSTARIALAGRSSER
MLBr_00129|M.leprae_Br4923 MSPDK---REFDIVLYGATGFSGKLTAEHLAL--SESTARIALAGRSSER
MMAR_1757|M.marinum_M MSSAQ---REFDIVLYGATGFSGMLTGQHLAQ--RDTNARIALAGRSPQR
MUL_2000|M.ulcerans_Agy99 MSSAQ---REFEIVLYGATGFSGMLTGQHLAQ--RDTNARIALAGRSPQR
* *.::*:*****: * **. :** ******** ::
Mflv_2628|M.gilvum_PYR-GCK LLAVRDSLGEQAKDWPLIVADASQPSTINAMAASTRVVVTTVGPYLRYGL
Mvan_3953|M.vanbaalenii_PYR-1 LLAVRESLGEAAQDWPLLVADASQPSTLNAMAASTRVVITTVGPYLRYGL
TH_0608|M.thermoresistible__bu LAAVRESLGEAAASWPLLTADADQPATLDELASRTQVVVTTVGPYTKYGM
MSMEG_4632|M.smegmatis_MC2_155 LQAARDSIGGAAADWPLVIADADNPSTLADMAARTRVVVTTVGPYTRYGL
MAV_1723|M.avium_104 LHAVRDSLGDAAQSWPVLQADASSPATLNEMAARTQVVITTVGPYTRYGL
MAB_4339c|M.abscessus_ATCC_199 LAAVRDACGPAARDWPLIEADTTRPATLAAMAASTQVVLTTVGPYTKYGL
Rv2953|M.tuberculosis_H37Rv LRGVRMMLGPNAADWPLILADASQPLTLEAMAARAQVVLTTVGPYTRYGL
Mb2977|M.bovis_AF2122/97 LRGVRMMLGPNAADWPLILADASQPLTLEAMAARAQVVLTTVGPYTRYGL
MLBr_00129|M.leprae_Br4923 LRNVRALLGPNAQDWPLIVADASQPSTLEAMAGRAQVVLTTVGPYTRYGL
MMAR_1757|M.marinum_M LRAVRDKLGPLASDWPLVVADASQPATLEEMATRAQVILTTVGPYTRYGL
MUL_2000|M.ulcerans_Agy99 LRAVRDKLGPLASDWPLVVADASQPATLEEMATRAQVILTTVGPYTRYGL
* .* * * .**:: **: * *: :* ::*::****** :**:
Mflv_2628|M.gilvum_PYR-GCK PLVAACAAAGTDYADLTGETLFVRRAIDLHHKQAVDTGARIVHACGFDSI
Mvan_3953|M.vanbaalenii_PYR-1 PLVAACAAAGTDYADLTGETLFVRECIDLYHKQAADTGARIVHACGFDSI
TH_0608|M.thermoresistible__bu PLVAACAVAGTDYADLTGETMFIRRAIDLYHKQAADNGARIVHSCGFDSI
MSMEG_4632|M.smegmatis_MC2_155 PLVAACAAAGTDYADLTGETMFIRDSIDRYHQQAVDTGARIVHSCGFDSV
MAV_1723|M.avium_104 PLVAACAAAGTDYADLTGEAMFVRDSIDSYHKQAADTGARIVHACGFDSV
MAB_4339c|M.abscessus_ATCC_199 PLVAACAEAGTDYADLTGEVNFVRESIDVYGKQAADTGARIVHCCGFDSI
Rv2953|M.tuberculosis_H37Rv PLVAACAKAGTDYADLTGELMFCRNSIDLYHKQAADTGARIILACGFDSI
Mb2977|M.bovis_AF2122/97 PLVAACAKAGTDYADLTGELMFCRNSIDLYHKQAADTGARIILACGFDSI
MLBr_00129|M.leprae_Br4923 PLVAACARTGTDYADLTGELMFCRNSIDLHHKQAAATGARIILACGFDSV
MMAR_1757|M.marinum_M PLVAACAKAGTDYADLTGELMFCRNSIDLYHKQAADTGARIVLACGFDSI
MUL_2000|M.ulcerans_Agy99 PLVAACAKAGTDYADLTGELMFCRNSIDLYHKQAADTGARIVLACGFDSI
******* :********** * * .** : :**. .****: .*****:
Mflv_2628|M.gilvum_PYR-GCK PSDMTVFALYRQAERDGTGQLGDTNFVARSFAG-GVSGGTIASMTELARE
Mvan_3953|M.vanbaalenii_PYR-1 PSDMTVFALYRAAERDRTGELGDTNFVVRSFAG-GVSGGTVASMTELARQ
TH_0608|M.thermoresistible__bu PSDITVYALYRRAVEDGTGELGDTNLVVRRFAG-GVSGGTAASMLEVMRT
MSMEG_4632|M.smegmatis_MC2_155 PSDLTVFALHRRATADGAGELVDTDLVLRSFAG-GVSGGTAASMIEVLRT
MAV_1723|M.avium_104 PSDLSVYALYRAARDDGAGELVDTDLVVRSFSG-GVSGGTVASMLEVLDT
MAB_4339c|M.abscessus_ATCC_199 PSDLSVYALYRRAQEDGTGELLDTTYVLSKFRG-GVSGGTAASMVEVMEA
Rv2953|M.tuberculosis_H37Rv PSDLNVYQLYRRSVEDGTGELCDTDLVLRSFSQRWVSGGSVATYSEAMRT
Mb2977|M.bovis_AF2122/97 PSDLNVYQLYRRSVEDGTGELCDTDLVLRSFSQRWVSGGSVATYSEAMRT
MLBr_00129|M.leprae_Br4923 PSDLNVYQLYRRVIEDRTGELCDTDLVLRSFSQRWVSGGSVAAYSEAMLT
MMAR_1757|M.marinum_M PSDLNVHHLYRRAAEDGTGELAETNLVLRSFSQRWASGGSVATYSEAMRT
MUL_2000|M.ulcerans_Agy99 PSDLNVHHLYRRAAEDGTGELAETNLVLRSFSQRWASGGSVATYSEAMRT
***:.*. *:* * :*:* :* * * .***: *: *
Mflv_2628|M.gilvum_PYR-GCK ASLDPEARRLLNDPYTLTPDRGAEPELGPQPDARWRRGREIAPELDGYWV
Mvan_3953|M.vanbaalenii_PYR-1 ASQDPEARRLLNDPYTLTPDRAAEPELGAQPDARWRRGREIAPELDGYWV
TH_0608|M.thermoresistible__bu ASEDPEARRLMTDPYTLSPDRAAEPELGGQPDIRWRRGGEIAPELEGYWT
MSMEG_4632|M.smegmatis_MC2_155 SSTDPAARAAMTDPYTLSPDRSREPELGDQPDVRWRRGSEIAPELAGYWT
MAV_1723|M.avium_104 ASRDPEARRQLADPYTLSSDRGAEPDVGPQPDLPCRRGRQIAPELAGVWT
MAB_4339c|M.abscessus_ATCC_199 SAEDPEVRRNAQDPYSLSPDRPAEPEVGRQREFQTLRGESVAPELTGKWL
Rv2953|M.tuberculosis_H37Rv ASSDPEARRLVTDPYTLTTDRGAEPELGAQPDFLRRPGRDLAPELAGFWT
Mb2977|M.bovis_AF2122/97 ASSDPEARRLVTDPYTLTTDRGAEPELGAQPDFLRRPGRDLAPELAGFWT
MLBr_00129|M.leprae_Br4923 TSNDPEALRLVTDPYTLTTDRDAEPDLGPQPDFPRHRGSDLAPELAGFWT
MMAR_1757|M.marinum_M ASSDPEAYRLVNDPYTLTTDRSAEPDLGPQPDFSMRRGRDLAPELAGFWT
MUL_2000|M.ulcerans_Agy99 ASSDPEAYRLVNDPYTLTTDRSAEPDLGPQPDFSMRRGRDLAPELAGFWT
:: ** . ***:*:.** **::* * : * .:**** * *
Mflv_2628|M.gilvum_PYR-GCK GAFAMALPNTRVVRRSNALLDYAYGRRFEYAELMSTGRSIGAPVLAAVAT
Mvan_3953|M.vanbaalenii_PYR-1 GAFAMALPNTRVVRRSNALLGYAYGRRFEYAEQMSTGRSVGAPLVAAMAT
TH_0608|M.thermoresistible__bu GAFVMAVANTRIVRRTNALLDYAYGRRFEYAEQMSLGKSIAAPVAAALAT
MSMEG_4632|M.smegmatis_MC2_155 GAFAMAGPNTRIVRRSNALLDYAYGRRFAYAEQMSCGRSPAAPIAAALVT
MAV_1723|M.avium_104 AGFVMAPYNTRIVRRSNALLDWAYGRSLRYSESMSLGSSPLAPVASAVVG
MAB_4339c|M.abscessus_ATCC_199 GAFFMGPVNTRIVRRSNALLDWAYGTRMRYREAMSLGSSVVAPVAAAAVT
Rv2953|M.tuberculosis_H37Rv GGFVQAPFNTRIVRRSNALQEWAYGRRFRYSETMSLGKSMAAPILAAAVT
Mb2977|M.bovis_AF2122/97 GGFVQAPFNTRIVRRSNALQEWAYGRRFRYSETMSLGKSMAAPILAAAVT
MLBr_00129|M.leprae_Br4923 GGFVQAQFNTRIVRRSNALQNWSYGRQFRYSETMSLGKSWAAPVASAAVT
MMAR_1757|M.marinum_M GAFVQGPFNTRIVRRSNALQDWAYGRQFRYSETMSLGRSFAAPIASAAVT
MUL_2000|M.ulcerans_Agy99 GAFVQGPFNTRIVRRSNALQDWAYGKQFRYSETMSLGRSFAAPIASAAVT
..* . ***:***:*** ::** : * * ** * * **: :* .
Mflv_2628|M.gilvum_PYR-GCK AGNAATMQLGSRFLNRVPRAALERVLPKPGTGPNEQTRERGHYTVETYTT
Mvan_3953|M.vanbaalenii_PYR-1 AGNVATMELSSRFLDRVPRGALERILPKVGTGPSEQTRERGHYTVETYTT
TH_0608|M.thermoresistible__bu AGNAAAFGLGSRFFHRVPDRLIEKVMPKPGTGPSERTRERGHYTVETYTT
MSMEG_4632|M.smegmatis_MC2_155 ASNAATMGLGGRFFDKIPSGLLERVLPKPGTGPSEQTRERGHYRVETYTT
MAV_1723|M.avium_104 GTAAATFGLGSRYFRFLPRRLVERIVPKPGTGPSPSARERGYYRIETYTT
MAB_4339c|M.abscessus_ATCC_199 GALTVGFGLGTRVP--LPKFIAQRVLPKPGTGPSERTRERGHYRVETYTT
Rv2953|M.tuberculosis_H37Rv GTVAGTIGLGNKYFDRLPRRLVERVTPKPGTGPSRKTQERGHYTFETYTT
Mb2977|M.bovis_AF2122/97 GTVAGTIGLGNKYFDRLPRRLVERVTPKPGTGPSRKTQERGHYTFETYTT
MLBr_00129|M.leprae_Br4923 SVVAGAVGLGNKYFNRLPRRVVERVTPKSGTGPSRKTQARGHYTFETYTT
MMAR_1757|M.marinum_M GAIAGGIGLGNKYFSRLPQRALERVTPKPGTGPSQKSRERGHYRCETYTT
MUL_2000|M.ulcerans_Agy99 GAIAGGIGLGNKYFSRLPQRALERVTPKPGTGPSQKSRERGHYRCETYTT
. . . *. : :* ::: ** ****. :: **:* *****
Mflv_2628|M.gilvum_PYR-GCK TSTGARYVARMSQNGDPGYKATAVLLGESGLALALDRDKLSDLRGILTPA
Mvan_3953|M.vanbaalenii_PYR-1 TSTGARYLARMSQQGDPGYKATSVLLGESGLALALDRDKLSDLRGILTPA
TH_0608|M.thermoresistible__bu TTTGARYVARMSQQGDPGYQATSVLLGESGLALALDRDKLSDLRGVLTPA
MSMEG_4632|M.smegmatis_MC2_155 TTSGARYVATMAQQGDPGYKATAVLLGECGLALATDRDKLSGLHGVLTPA
MAV_1723|M.avium_104 TTSGARYVARMEQRGDPGYKATSVLLGESGLALAFDRDKLPQLYGVLTPA
MAB_4339c|M.abscessus_ATCC_199 TTEGVRYRAIIAQEGDPGYKATAVLLGESGLTLALDRTELPDRLGVLTPA
Rv2953|M.tuberculosis_H37Rv TTTGARYRATFAHNVD-AYKSTAVLLAQSGLALALDRDRLAELRGVLTPA
Mb2977|M.bovis_AF2122/97 TTTGARYRATFAHNVD-AYKSTAVLLAQSGLALALDRDRLAELRGVLTPA
MLBr_00129|M.leprae_Br4923 TTTGARYMATFAHNVD-AYKSTAGLLAASGLTLALDRDRLSELRGVLTPA
MMAR_1757|M.marinum_M TTTGARYLATFAHNVD-AYASTAVLLGESGLALALDRDRLSELRGVLTPA
MUL_2000|M.ulcerans_Agy99 TTTGARYLATFAHNVD-AYASTAVLLGESGLALALDRDRLSELRGVLTPA
*: *.** * : :. * .* :*: **. .**:** ** .*. *:****
Mflv_2628|M.gilvum_PYR-GCK AAMGDALLARCPGA-DVSLDVSRLN-
Mvan_3953|M.vanbaalenii_PYR-1 AAMGDALLARFPAA-GVSLDVSRLN-
TH_0608|M.thermoresistible__bu AAMGDVLLTRLPAA-GVTLTVERLA-
MSMEG_4632|M.smegmatis_MC2_155 AAMGDALLTRLPAA-GVVLETTKLD-
MAV_1723|M.avium_104 AAMGDALLDRFPGA-GVSLQVDRLAG
MAB_4339c|M.abscessus_ATCC_199 AAMGDPLLERLRAARGVTVEVERL--
Rv2953|M.tuberculosis_H37Rv AAMGDALLARLPGA-GVVMGTTRLS-
Mb2977|M.bovis_AF2122/97 AAMGDALLARLPGA-GVVMGTTRLS-
MLBr_00129|M.leprae_Br4923 AAMGEALLTRLPSA-GVVIGTTRLS-
MMAR_1757|M.marinum_M AAMGDALLARLPQT-GVVVSATRLH-
MUL_2000|M.ulcerans_Agy99 AAMGDALLARLPQT-GVVVSATRLH-
****: ** * : .* : . :*