For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQTPDREKALELAVAQIEKSYGKGSVMRLGDEARQPISVIPTGSIALDVALGIGGLPRGRVIEIYGPESS GKTTVALHAVANAQAAGGVAAFIDAEHALDPDYAKKLGVDTDSLLVSQPDTGEQALEIADMLIRSGALDI VVIDSVAALVPRAELEGEMGDSHVGLQARLMSQALRKMTGALNNSGTTAIFINQLRDKIGVMFGSPETTT GGKALKFYASVRMDVRRVETLKDGTNAVGNRTRVKVVKNKCLAEGTRIFDPVTGTTHRIEDVVDGRKPIH VVAAAKDGTLHARPVVSWFDQGTRDVIGLRIAGGAIVWATPDHKVLTEYGWRAAGELRKGDRVAQPRRFD GFGDSAPIPADHARLLGYLIGDGRDGWVGGKTPINFINVQRALIDDVTRIAATLGCAAHPQGRISLAIAH RPGERNGVADLCQQAGIYGKLAWEKTIPNWFFEPDIAADIVGNLLFGLFESDGWVSREQTGALRVGYTTT SEQLAHQIHWLLLRFGVGSTVRDYDPTQKRPSIVNGRRIQSKRQVFEVRISGMDNVTAFAESVPMWGPRG AALIQAIPEATQGRRRGSQATYLAAEMTDAVLNYLDERGVTAQEAAAMIGVASGDPRGGMKQVLGASRLR RDRVQALADALDDKFLHDMLAEELRYSVIREVLPTRRARTFDLEVEELHTLVAEGVVVHNCSPPFKQAEF DILYGKGISREGSLIDMGVDQGLIRKSGAWFTYEGEQLGQGKENARNFLVENADVADEIEKKIKEKLGIG AVVTDDPSNDGVLPAPVDF*
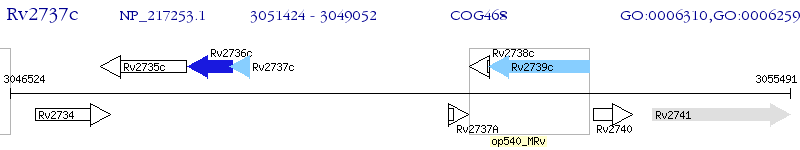
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2737c | recA | - | 100% (790) | recombinase A |
| M. tuberculosis H37Rv | Rv0058 | dnaB | 2e-19 | 25.33% (450) | replicative DNA helicase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2756c | recA | 0.0 | 100.00% (790) | DNA recombination protein RecA |
| M. gilvum PYR-GCK | Mflv_3975 | recA | 1e-128 | 92.37% (249) | recA protein |
| M. leprae Br4923 | MLBr_00987 | recA | 1e-109 | 61.88% (383) | RecA protein |
| M. abscessus ATCC 19977 | MAB_3060c | - | 1e-130 | 92.43% (251) | RecA protein |
| M. marinum M | MMAR_1977 | recA | 1e-134 | 96.81% (251) | recombinase A RecA |
| M. avium 104 | MAV_3627 | recA | 1e-133 | 96.41% (251) | protein RecA |
| M. smegmatis MC2 155 | MSMEG_2723 | recA | 1e-129 | 93.17% (249) | protein RecA |
| M. thermoresistible (build 8) | TH_1955 | recA | 1e-112 | 60.84% (383) | RECA PROTEIN (RECOMBINASE A) |
| M. ulcerans Agy99 | MUL_3380 | recA | 1e-133 | 96.41% (251) | recombinase A |
| M. vanbaalenii PYR-1 | Mvan_2420 | recA | 1e-129 | 93.17% (249) | recA protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3975|M.gilvum_PYR-GCK MAQQAPDREKALELALAQIDKNFGKGSVMRLGEDVRPPIAVIPTGSIALD
Mvan_2420|M.vanbaalenii_PYR-1 MAQQAPDREKALELALAQIDKNFGKGSVMRLGEEVRQPIAVIPTGSIALD
MAB_3060c|M.abscessus_ATCC_199 -MAQAPDREKALELALAQIDKSFGKGSVMRLGDEVRQPISVIPTGSIALD
TH_1955|M.thermoresistible__bu -MAQAPDREKALELALAQIEKNHGKGSVMRLGDEARQPISVIPTGSIALD
MLBr_00987|M.leprae_Br4923 -MAQVPDREKALELAMAQIEKNYGKGSVMRLGDEMCQPISVIPTGSIALD
MSMEG_2723|M.smegmatis_MC2_155 MAQQAPDREKALELAMAQIDKNFGKGSVMRLGEEVRQPISVIPTGSISLD
MAV_3627|M.avium_104 -MTQAPDREKALELAMAQIEKSYGKGSVMRLGDEMRQPISVIPTGSIALD
Rv2737c|M.tuberculosis_H37Rv -MTQTPDREKALELAVAQIEKSYGKGSVMRLGDEARQPISVIPTGSIALD
Mb2756c|M.bovis_AF2122/97 -MTQTPDREKALELAVAQIEKSYGKGSVMRLGDEARQPISVIPTGSIALD
MUL_3380|M.ulcerans_Agy99 -MAQAPDREKALELAMAQIEKSYGKGSVMRLGDEVRQPISIIPTGSIALD
MMAR_1977|M.marinum_M -MAQAPDREKALELAMAQIEKSYGKGSVMRLGDEVRQPISIIPTGSIALD
*.**********:***:*..*********:: **::******:**
Mflv_3975|M.gilvum_PYR-GCK VALGIGGLPRGRVIEIYGPESSGKTTVALHAVANAQAAGGIAAFIDAEHA
Mvan_2420|M.vanbaalenii_PYR-1 VALGIGGLPRGRVVEIYGPESSGKTTVALHAVANAQAAGGIAAFIDAEHA
MAB_3060c|M.abscessus_ATCC_199 VALGIGGLPRGRVIEIYGPESSGKTTVALHAVANAQRAGGIAAFIDAEHA
TH_1955|M.thermoresistible__bu VALGIGGLPRGRVVEIYGPESSGKTTVALHAVANAQAAGGIAAFIDAEHA
MLBr_00987|M.leprae_Br4923 VALGIGGLPRGRIVEIYGPESSGKTTVALHAVANAQAVGGVAAFIDAEHA
MSMEG_2723|M.smegmatis_MC2_155 VALGIGGLPRGRVIEIYGPESSGKTTVALHAVANAQAAGGIAAFIDAEHA
MAV_3627|M.avium_104 VALGIGGLPRGRVVEIYGPESSGKTTVALHAVANAQAAGGVAAFIDAEHA
Rv2737c|M.tuberculosis_H37Rv VALGIGGLPRGRVIEIYGPESSGKTTVALHAVANAQAAGGVAAFIDAEHA
Mb2756c|M.bovis_AF2122/97 VALGIGGLPRGRVIEIYGPESSGKTTVALHAVANAQAAGGVAAFIDAEHA
MUL_3380|M.ulcerans_Agy99 VALGIGGLPRGRVIEIYGPESSGKTTVALHAVANAQAAGGVAAFIDAEHA
MMAR_1977|M.marinum_M VALGIGGLPRGRVIEIYGPESSGKTTVALHAVANAQAAGGVAAFIDAEHA
************::********************** .**:*********
Mflv_3975|M.gilvum_PYR-GCK LDPDYAKKLGVDTDALLVSQPDTGEQALEIADMLIRSGALDILVIDSVAA
Mvan_2420|M.vanbaalenii_PYR-1 LDPDYAQKLGVDTDALLVSQPDTGEQALEIADMLIRSGALDILVIDSVAA
MAB_3060c|M.abscessus_ATCC_199 LDPEYAKKLGVDTDSLLVSQPDTGEQALEIADMLVRSGAIDLIVIDSVAA
TH_1955|M.thermoresistible__bu LDPEYAKKLGVDTDALLVSQPDTGEQALEIADMLIRSGALDILVVDSVAA
MLBr_00987|M.leprae_Br4923 LEPEYAKKLGVDTDSLLVSQPDTGEQALEIADMLIRSGALDIVVIDSVAA
MSMEG_2723|M.smegmatis_MC2_155 LDPEYAKKLGVDTDSLLVSQPDTGEQALEIADMLVRSGALDIIVIDSVAA
MAV_3627|M.avium_104 LDPEYAKKLGVDTDSLLVSQPDTGEQALEIADMLIRSGALDILVIDSVAA
Rv2737c|M.tuberculosis_H37Rv LDPDYAKKLGVDTDSLLVSQPDTGEQALEIADMLIRSGALDIVVIDSVAA
Mb2756c|M.bovis_AF2122/97 LDPDYAKKLGVDTDSLLVSQPDTGEQALEIADMLIRSGALDIVVIDSVAA
MUL_3380|M.ulcerans_Agy99 LDPEYAKKLGVDTDSLLVSQPDTGEQALEIVDMLIRSGALDIVVIDSVAA
MMAR_1977|M.marinum_M LDPEYAKKLGVDTDSLLVSQPDTGEQALEIADMLIRSGALDIVVIDSVAA
*:*:**:*******:***************.***:****:*::*:*****
Mflv_3975|M.gilvum_PYR-GCK LVPRAEIEGEMGDSHVGLQARLMSQALRKMTGALNNSGTTAIFINQLREK
Mvan_2420|M.vanbaalenii_PYR-1 LVPRAEIEGEMGDSHVGLQARLMSQALRKMTGALNNSGTTAIFINQLREK
MAB_3060c|M.abscessus_ATCC_199 LVPRAEIEGEMGDSHVGLQARLMSQALRKMTGALNNSGTTAIFINQLREK
TH_1955|M.thermoresistible__bu LVPRAEIEGEMGDSHVGLQARLMSQALRKITGALSNSGTTAIFINQLREK
MLBr_00987|M.leprae_Br4923 LVPRAELEGEMGDSYVGLQARLMSQALRKMTGALSNSGTTAIFINQLREK
MSMEG_2723|M.smegmatis_MC2_155 LVPRAEIEGEMGDSHVGLQARLMSQALRKMTGALNNSGTTAIFINQLREK
MAV_3627|M.avium_104 LVPRAELEGEMGDSHVGLQARLMSQALRKMTGALNNSGTTAIFINQLREK
Rv2737c|M.tuberculosis_H37Rv LVPRAELEGEMGDSHVGLQARLMSQALRKMTGALNNSGTTAIFINQLRDK
Mb2756c|M.bovis_AF2122/97 LVPRAELEGEMGDSHVGLQARLMSQALRKMTGALNNSGTTAIFINQLRDK
MUL_3380|M.ulcerans_Agy99 LVPRAELEGEMGDSHVGLQARLMSQALRKMTGALNNSGTTAIFINQLRDK
MMAR_1977|M.marinum_M LVPRAELEGEMGDSHVGLQARLMSQALRKMTGALNNSGTTAIFINQLRDK
******:*******:**************:****.*************:*
Mflv_3975|M.gilvum_PYR-GCK IGVMFG--------------------------------------------
Mvan_2420|M.vanbaalenii_PYR-1 IGVMFG--------------------------------------------
MAB_3060c|M.abscessus_ATCC_199 IGVMFG--------------------------------------------
TH_1955|M.thermoresistible__bu IGVMFGCFDYST--------------------------------RVQLAD
MLBr_00987|M.leprae_Br4923 IGVMFGCMNYST--------------------------------RVTLAD
MSMEG_2723|M.smegmatis_MC2_155 IGVMFG--------------------------------------------
MAV_3627|M.avium_104 IGVMFG--------------------------------------------
Rv2737c|M.tuberculosis_H37Rv IGVMFGSPETTTGGKALKFYASVRMDVRRVETLKDGTNAVGNRTRVKVVK
Mb2756c|M.bovis_AF2122/97 IGVMFGSPETTTGGKALKFYASVRMDVRRVETLKDGTNAVGNRTRVKVVK
MUL_3380|M.ulcerans_Agy99 IGVMFGSPETTTGGKALKFYASVRMDVRRIETLKDGTNAVGNRTRVKIVK
MMAR_1977|M.marinum_M IGVMFGSPETTTGGKALKFYASVRMDVRRIETLKDGTNAVGNRTRVKIVK
******
Mflv_3975|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2420|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3060c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1955|M.thermoresistible__bu GTTEKIGKIVDNKMDVEVLSYDPDTDRIVPRKVVNWFNNGPAEQFLQFTV
MLBr_00987|M.leprae_Br4923 GSTEKIGKIVNNKMDVRVLSYDPVTDRIVPRKVVNWFNNGPAEQFLQFTV
MSMEG_2723|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3627|M.avium_104 --------------------------------------------------
Rv2737c|M.tuberculosis_H37Rv NKCLAEGTRIFDPVTGTTHRIEDVVDGRKPIHVVAAAKDGTLHARPVVSW
Mb2756c|M.bovis_AF2122/97 NKCLAEGTRIFDPVTGTTHRIEDVVDGRKPIHVVAAAKDGTLHARPVVSW
MUL_3380|M.ulcerans_Agy99 N-------------------------------------------------
MMAR_1977|M.marinum_M N-------------------------------------------------
Mflv_3975|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2420|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3060c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1955|M.thermoresistible__bu EKSGGN--------GRARFAATPNHLIRTPGGWTEAGDLIAGDRVLAAEP
MLBr_00987|M.leprae_Br4923 EKSGSN--------GKSQFAATPNHLIRTPGGWTEAGNLIAGDRVLAVEP
MSMEG_2723|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3627|M.avium_104 --------------------------------------------------
Rv2737c|M.tuberculosis_H37Rv FDQGTRDVIGLRIAGGAIVWATPDHKVLTEYGWRAAGELRKGDRVAQPRR
Mb2756c|M.bovis_AF2122/97 FDQGTRDVIGLRIAGGAIVWATPDHKVLTEYGWRAAGELRKGDRVAQPRR
MUL_3380|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1977|M.marinum_M --------------------------------------------------
Mflv_3975|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2420|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3060c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1955|M.thermoresistible__bu HRLSDQQFQIVLGSLMGDGTLSPDPRGRNGVRFRMG-HGADRVDYLEWKT
MLBr_00987|M.leprae_Br4923 HMLSDQQFQVVLGSLMGDGNLSPNLCDRNGVRFRLLGYGCKQVEYLQWKK
MSMEG_2723|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3627|M.avium_104 --------------------------------------------------
Rv2737c|M.tuberculosis_H37Rv FDGFGDSAPIPADHARLLGYLIGDGRDGWVGGKTPINFINVQRALIDDVT
Mb2756c|M.bovis_AF2122/97 FDGFGDSAPIPADHARLLGYLIGDGRDGWVGGKTPINFINVQRALIDDVT
MUL_3380|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1977|M.marinum_M --------------------------------------------------
Mflv_3975|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2420|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3060c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1955|M.thermoresistible__bu ALLGNIKHSTGENAEGARFVDFTPLPELAELRRAVYLGDDGRKFISEEYL
MLBr_00987|M.leprae_Br4923 ALMGNIRHTVRENSMGASFIDFTPLPELVELQRAVYLGD-GKKFLSEEYL
MSMEG_2723|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3627|M.avium_104 --------------------------------------------------
Rv2737c|M.tuberculosis_H37Rv RIAATLGCAAHPQGRISLAIAHRPGERNGVADLCQQAGIYGKLAWEKTIP
Mb2756c|M.bovis_AF2122/97 RIAATLGCAAHPQGRISLAIAHRPGERNGVADLCQQAGIYGKLAWEKTIP
MUL_3380|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1977|M.marinum_M --------------------------------------------------
Mflv_3975|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2420|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3060c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1955|M.thermoresistible__bu KALTPLALAIWYMDDGSLTV-RSEGLQQGTAGGSGRIEICVEAMTEGSRI
MLBr_00987|M.leprae_Br4923 KALTPLVLAIWYMDDGSFTV-GSKRVQERTAGGSGRIEICVDAMTEGTRV
MSMEG_2723|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3627|M.avium_104 --------------------------------------------------
Rv2737c|M.tuberculosis_H37Rv NWFFEPDIAADIVGNLLFGLFESDGWVSREQTGALRVGYTTTSEQLAHQI
Mb2756c|M.bovis_AF2122/97 NWFFEPDIAADIVGNLLFGLFESDGWVSREQTGALRVGYTTTSEQLAHQI
MUL_3380|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1977|M.marinum_M --------------------------------------------------
Mflv_3975|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2420|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3060c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1955|M.thermoresistible__bu RLRDHLRDTHGLDVRLRQAGAGGKAVLVFSTAATAEFQELVAPYMAPSME
MLBr_00987|M.leprae_Br4923 RLRDYLCDTHGLDVRLREVGSAGKAVLVFSTAATAKFQSLIAPYVAPSME
MSMEG_2723|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3627|M.avium_104 --------------------------------------------------
Rv2737c|M.tuberculosis_H37Rv HWLLLRFGVGSTVRDYDPTQKRPSIVNGRRIQSKRQVFEVRISGMDNVTA
Mb2756c|M.bovis_AF2122/97 HWLLLRFGVGSTVRDYDPTQKRPSIVNGRRIQSKRQVFEVRISGMDNVTA
MUL_3380|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1977|M.marinum_M --------------------------------------------------
Mflv_3975|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2420|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3060c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1955|M.thermoresistible__bu YKLLPRFRGQSRVVPQFVEPTQRLVPARILDVHVEPHTRSMNRYDIEVEG
MLBr_00987|M.leprae_Br4923 YKLLPQFRGRGSVTPQFVEPTQQLVPARVLDVHVKLSTRSMNRFDIEVEG
MSMEG_2723|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3627|M.avium_104 --------------------------------------------------
Rv2737c|M.tuberculosis_H37Rv FAESVPMWGPRGAALIQAIPEATQGRRRGSQATYLAAEMTDAVLNYLDER
Mb2756c|M.bovis_AF2122/97 FAESVPMWGPRGAALIQAIPEATQGRRRGSQATYLAAEMTDAVLNYLDER
MUL_3380|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1977|M.marinum_M --------------------------------------------------
Mflv_3975|M.gilvum_PYR-GCK -------------SPETTTGG---------------KALK----------
Mvan_2420|M.vanbaalenii_PYR-1 -------------SPETTTGG---------------KALK----------
MAB_3060c|M.abscessus_ATCC_199 -------------SPETTTGG---------------KALK----------
TH_1955|M.thermoresistible__bu NHNYFVDGVMVHNSPETTTGG---------------KALK----------
MLBr_00987|M.leprae_Br4923 NHNYFVDGVMVHNSPETTTGG---------------KALK----------
MSMEG_2723|M.smegmatis_MC2_155 -------------SPETTTGG---------------KALK----------
MAV_3627|M.avium_104 -------------SPETTTGG---------------KALK----------
Rv2737c|M.tuberculosis_H37Rv GVTAQEAAAMIGVASGDPRGGMKQVLGASRLRRDRVQALADALDDKFLHD
Mb2756c|M.bovis_AF2122/97 GVTAQEAAAMIGVASGDPRGGMKQVLGASRLRRDRVQALADALDDKFLHD
MUL_3380|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1977|M.marinum_M --------------------------------------------------
Mflv_3975|M.gilvum_PYR-GCK ------FYSSVR--LDVRRIETLKDGTDAVGNRTRVKVVKNKVSPPFKQA
Mvan_2420|M.vanbaalenii_PYR-1 ------FYASVR--MDVRRIETLKDGTDAVGNRTRVKVVKNKVSPPFKQA
MAB_3060c|M.abscessus_ATCC_199 ------FYSSVR--LDVRRIETLKDGTDAVGNRTRVKVVKNKVSPPFKQA
TH_1955|M.thermoresistible__bu ------FYASVR--IDVRRIETLKDGTEAVGNRTRAKIVKNKVSPPFKQA
MLBr_00987|M.leprae_Br4923 ------FYASVR--MDVRRIETLKDGVDAVGNRTRVKIVKNKVSPPFKQA
MSMEG_2723|M.smegmatis_MC2_155 ------FYASVR--LDVRRIETLKDGTDAVGNRTRVKVVKNKVSPPFKQA
MAV_3627|M.avium_104 ------FYASVR--MDVRRIETLKDGTNAVGNRTRVKIVKNKVSPPFKQA
Rv2737c|M.tuberculosis_H37Rv MLAEELRYSVIREVLPTRRARTFDLEVEELHTLVAEGVVVHNCSPPFKQA
Mb2756c|M.bovis_AF2122/97 MLAEELRYSVIREVLPTRRARTFDLEVEELHTLVAEGVVVHNCSPPFKQA
MUL_3380|M.ulcerans_Agy99 -----------------------------------------KVSPPFKQA
MMAR_1977|M.marinum_M -----------------------------------------KVSPPFKQA
: *******
Mflv_3975|M.gilvum_PYR-GCK EFDILYGKGISREGSLIDMGVEQGFIRKSGSWFTYDGEQLGQGKENARNF
Mvan_2420|M.vanbaalenii_PYR-1 EFDILYGKGISREGSLIDMGVEHGFIRKSGSWFTYDGEQLGQGKENARNF
MAB_3060c|M.abscessus_ATCC_199 EFDILYGKGISKEGSLIDMGVEQGFIRKSGSWFTYEGEQLGQGKENARNF
TH_1955|M.thermoresistible__bu EFDILYGKGISKEGSLIDMGVEQGFIRKSGSWYTYEGEQLGQGKENARNF
MLBr_00987|M.leprae_Br4923 EFDILYGKGISREGSLIDMGVEQGFVRKSGSWFTYEGEQLGQGKENARNF
MSMEG_2723|M.smegmatis_MC2_155 EFDILYGQGISREGSLIDMGVEHGFIRKSGSWFTYEGEQLGQGKENARKF
MAV_3627|M.avium_104 EFDILYGRGISREGSLIDMGVDQGFIRKSGSWFTYEGEQLGQGKENARTF
Rv2737c|M.tuberculosis_H37Rv EFDILYGKGISREGSLIDMGVDQGLIRKSGAWFTYEGEQLGQGKENARNF
Mb2756c|M.bovis_AF2122/97 EFDILYGKGISREGSLIDMGVDQGLIRKSGAWFTYEGEQLGQGKENARNF
MUL_3380|M.ulcerans_Agy99 EFDILYGRGISREGSLIDMGVDQGFIRKSGAWFTYEGEQLGQGKENARNF
MMAR_1977|M.marinum_M EFDILYGRGISREGSLIDMGVDQGFIRKSGAWFTYEGEQLGQGKENARNF
*******:***:*********::*::****:*:**:************.*
Mflv_3975|M.gilvum_PYR-GCK LLQNPDVGNEIEKKIKEKLGIGAQLTDDVA-DDALPAPVDF
Mvan_2420|M.vanbaalenii_PYR-1 LLQNVDVANEIEKKIKEKLGIGAQLTDDAN-DDALPAPVDF
MAB_3060c|M.abscessus_ATCC_199 LLENVDVANEIEKKIKEKLGIGAVLTD----DEVVPAPVDF
TH_1955|M.thermoresistible__bu LIENVDVANEIEKKIKEKLGIGPVITD----DEALPAPVDF
MLBr_00987|M.leprae_Br4923 LLENADVANEIEKKIKEKLGIGAVVTD----DDILPTPVDF
MSMEG_2723|M.smegmatis_MC2_155 LLENTDVANEIEKKIKEKLGIGAVVTAEA--DDVLPAPVDF
MAV_3627|M.avium_104 LMENDEVANEIEKKIKEKLGIGAVVTDDLSDDGVLPAPVDF
Rv2737c|M.tuberculosis_H37Rv LVENADVADEIEKKIKEKLGIGAVVTDDPSNDGVLPAPVDF
Mb2756c|M.bovis_AF2122/97 LVENADVADEIEKKIKEKLGIGAVVTDDPSNDGVLPAPVDF
MUL_3380|M.ulcerans_Agy99 LLENGGVANEIEKKIKEKLGIGAVVTDD----GVLPAPVDF
MMAR_1977|M.marinum_M LLENGEVANEIEKKIKEKLGIGAVVTDD----GVLPAPVDF
*::* *.:*************. :* :*:****