For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNTATRVRLARKRADRLNLKLIKNGHHFRLRDADEITLAVGHLGVVEAFLAAAKSQNKPPGPPPSLHAPP SWRRDIDDYLLNLNAAGQRPATIRLRKTVLCAAAHGLGRPPADVTAEHLLDWLGKQQHLSPEGRKTYRST LRGFFVWAYEMDRVRDYVADSLPKVRCPKQPPRPAGDDVWQAALAKADRRIELMIRLAGEAGLRRAEAAQ AHTGDLMDGGLLLVHGKGGKRRIVPISDYLAALIRDTPHGYLFPNGTGGHLTAEHVGKLVSRALPGDATM HTLRHRYATRAYRGSHNLRAVQQLLGHASIVTTERYTALCDDEVRAAAAAAW
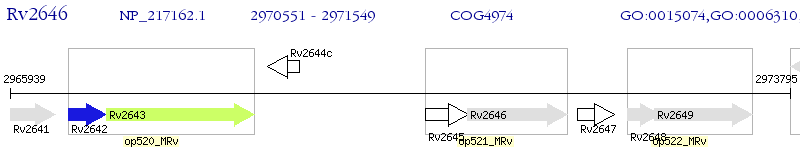
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2646 | - | - | 100% (332) | PROBABLE INTEGRASE |
| M. tuberculosis H37Rv | Rv1701 | xerD | 2e-12 | 30.43% (161) | site-specific tyrosine recombinase XerD |
| M. tuberculosis H37Rv | Rv2894c | xerC | 8e-10 | 28.96% (221) | site-specific tyrosine recombinase XerC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1727 | xerD | 2e-12 | 30.43% (161) | site-specific tyrosine recombinase XerD |
| M. gilvum PYR-GCK | Mflv_3500 | xerD | 4e-12 | 30.67% (163) | site-specific tyrosine recombinase XerD |
| M. leprae Br4923 | MLBr_01365 | xerD | 8e-12 | 29.52% (166) | site-specific tyrosine recombinase XerD |
| M. abscessus ATCC 19977 | MAB_2366 | xerD | 6e-14 | 31.71% (164) | site-specific tyrosine recombinase XerD |
| M. marinum M | MMAR_2506 | xerD | 1e-12 | 30.43% (161) | integrase/recombinase, XerD |
| M. avium 104 | MAV_3070 | xerD | 3e-12 | 30.67% (163) | site-specific tyrosine recombinase XerD |
| M. smegmatis MC2 155 | MSMEG_3744 | xerD | 1e-13 | 32.30% (161) | site-specific tyrosine recombinase XerD |
| M. thermoresistible (build 8) | TH_2517 | - | 4e-18 | 29.61% (233) | PUTATIVE putative site-specific recombinase, phage integrase |
| M. ulcerans Agy99 | MUL_1690 | xerD | 1e-12 | 30.43% (161) | site-specific tyrosine recombinase XerD |
| M. vanbaalenii PYR-1 | Mvan_3281 | xerD | 2e-12 | 31.29% (163) | site-specific tyrosine recombinase XerD |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2506|M.marinum_M -----MSTLTLDTQLQGYLDHLAIERG------------VAANTLSSYRR
MUL_1690|M.ulcerans_Agy99 -----MSTLTLDTQLQGYLDHLAIERG------------VAANTLSSYRR
Mb1727|M.bovis_AF2122/97 -----MKTLAL--QLQGYLDHLTIERG------------VAANTLSSYRR
MLBr_01365|M.leprae_Br4923 -----MTTAALQTQLQGYLDYLIIERS------------IAANTLSSYRR
MAV_3070|M.avium_104 -----MTTVALETQLQGYLDHLTIERG------------VAANTLSSYRR
Mflv_3500|M.gilvum_PYR-GCK ---MGVG-AVLDGQLQGYLDHLAIERG------------VAANTLSSYRR
Mvan_3281|M.vanbaalenii_PYR-1 MT-TGVDPAALDDQFQGYLDHLAIERG------------VAANTLSSYRR
MSMEG_3744|M.smegmatis_MC2_155 MTGTSALRAVLDEQVQGYLDHLTIERG------------VAANTLSSYRR
MAB_2366|M.abscessus_ATCC_1997 ---MTAAVGLLDGEIQSYLDHLDVERG------------VAANTLSSYRR
Rv2646|M.tuberculosis_H37Rv --MNTATRVRLARKRADRLNLKLIKNGHHFRLRDADEITLAVGHLGVVEA
TH_2517|M.thermoresistible__bu -------LESVQYPLVNTWRTWQFAQS------------LSKRTVDERVA
: . . .. :: :.
MMAR_2506|M.marinum_M DLRRYSKHLEDRGITDLAKVG---EDDVSEFLVALRRGDPESGVLGLSAV
MUL_1690|M.ulcerans_Agy99 DLRRYSKHLEDRGITDLAKVG---EDDVSEFLVALRRGDPESGVLGLSAV
Mb1727|M.bovis_AF2122/97 DLRRYSKHLEERGITDLAKVG---EHDVSEFLVALRRGDPDSGTAALSAV
MLBr_01365|M.leprae_Br4923 DLIRYSKHLSDRGIEDLAKVG---EHDVSEFLVALRRGDPDSGVAALSAV
MAV_3070|M.avium_104 DLRRYTKHLSDRGISDLAKVG---EDDVSDFLVALRRGDPDTGAAALSAV
Mflv_3500|M.gilvum_PYR-GCK DLRRYAEHLKARGVGDLAAVT---ETDVSEFLVALRLGDPDGGVVPLSAV
Mvan_3281|M.vanbaalenii_PYR-1 DLRRYAEHLASRGVTDLAGVA---EADVSEFLVSLRRGDPDNGVVPLSAV
MSMEG_3744|M.smegmatis_MC2_155 DLRRYAEHLRQRGIDDLARVA---ESDVSDFLVALRRGDPDAGVTALSAV
MAB_2366|M.abscessus_ATCC_1997 DLRRYQQHLADRKIDRLAEVC---ETDVSDFVVTLRRGDPANGVPELSAS
Rv2646|M.tuberculosis_H37Rv FLAAAKSQNKPPGPPPSLHAPPSWRRDIDDYLLNLNAAGQRPATIRLRKT
TH_2517|M.thermoresistible__bu TVTRMANWCQTSP-----------ESATSEQIAEWLAAGDWMPRTRWTYH
: . . .: : ..
MMAR_2506|M.marinum_M SAARALIAVRGLHRFAAAEGLAALDVARAVRPPTPGRRLPKSLT------
MUL_1690|M.ulcerans_Agy99 SAARALIAVRGLHRFAAAEGLAALDVARAVRPPTPGRRLPKSLT------
Mb1727|M.bovis_AF2122/97 SAARALIAVRGLHRFAAAEGLAELDVARAVRPPTPSRRLPKSLT------
MLBr_01365|M.leprae_Br4923 SAARALIAVRGLHRFAVAEGLVDLDVARAVRPPTPGRRLPKSLT------
MAV_3070|M.avium_104 SAARALIAVRGLHRFLAAEGLAELDVARAVRPPTPGRRLPKSLT------
Mflv_3500|M.gilvum_PYR-GCK SAARALIAVRGLHRFAAAEGIAAIDVARDVKPPTPSRRLPKSLS------
Mvan_3281|M.vanbaalenii_PYR-1 SAARAVIAVRGLHRFAAAEGITTVDVAREVKPPTPGRRLPKSLS------
MSMEG_3744|M.smegmatis_MC2_155 SAARAVVAVRGLHRFAAAEGLTENDVARAVKPPTPSRRLPKSLT------
MAB_2366|M.abscessus_ATCC_1997 SAARALIAVRGLHRFAAIEGLAPTDVARAVKPPTPNRRLPKSLT------
Rv2646|M.tuberculosis_H37Rv VLCAAAHGLGRPPADVTAEHLLDWLGKQQHLSPEGRKTYRSTLRGFFVWA
TH_2517|M.thermoresistible__bu RALSAWFTWLQLQGHRLDNPMLRLG------DPKRPKSVPRPLS------
* : : * : .*
MMAR_2506|M.marinum_M --IDEVLALLEG-------------AGGDNPADGPLTLRNR---ALLELL
MUL_1690|M.ulcerans_Agy99 --IDEVLALLEG-------------AGGDNPADGPLTLRNR---ALLELL
Mb1727|M.bovis_AF2122/97 --IDEVLSLLEG-------------AGGDKPSDGPLTLRNR---AVLELL
MLBr_01365|M.leprae_Br4923 --VDEVLALLES-------------VGGESRADGPLVLRNR---ALLELL
MAV_3070|M.avium_104 --IDQVLALLEA-------------AGGESPADGPLTLRNR---ALLELL
Mflv_3500|M.gilvum_PYR-GCK --IDEVLALLEA-------------AGGDSEADSPLTLRNR---ALLELL
Mvan_3281|M.vanbaalenii_PYR-1 --IDEVLALLEA-------------AGGDGEADGPLTLRNR---ALLELL
MSMEG_3744|M.smegmatis_MC2_155 --LDEVLALLEA-------------AGGDSEADSPLTLRNR---ALLELL
MAB_2366|M.abscessus_ATCC_1997 --VEQVEALLNA-------------AGGGG-SDGPLDLRNR---ALLELL
Rv2646|M.tuberculosis_H37Rv YEMDRVRDYVADSLPKVRCPKQPPRPAGDDVWQAALAKADRRIELMIRLA
TH_2517|M.thermoresistible__bu --NADVARLLTT----------------------RMHTRTR---AMVLLA
* : : * :: *
MMAR_2506|M.marinum_M YSTGSRISEAVGLDVDDIDTQARTVLLQGKGGKQRLVPVGRPAVQALDAY
MUL_1690|M.ulcerans_Agy99 YSTGSRISEAVGLDVDDIDTQARTVLLQGKGGKQRLVPVGRPAVQALDAY
Mb1727|M.bovis_AF2122/97 YSTGARISEAVGLDLDDIDTHARSVLLRGKGGKQRLVPVGRPAVHALDAY
MLBr_01365|M.leprae_Br4923 YSTGSRISEAVGLDVDDVDTQARTVLLQGKGGKQRLVPVGRPAVQALDAY
MAV_3070|M.avium_104 YSTGARISEAVGLDVDDVDTQARSVLLRGKGGKQRLVPIGRPAVAALDAY
Mflv_3500|M.gilvum_PYR-GCK YSTGARISEAVGLDIDDVDTSARSVLLRGKGGKQRLIPIGRPAVTALETY
Mvan_3281|M.vanbaalenii_PYR-1 YSTGARISEAVGLDVDDVDTEARSVLLRGKGGKQRLIPIGRPAVSALDNY
MSMEG_3744|M.smegmatis_MC2_155 YSTGARISEAVGLDVDDVDTQARSVLLHGKGGKQRLVPIGRPAVSALDAY
MAB_2366|M.abscessus_ATCC_1997 YSTGARISEAVGLDVDDVDVQARSVLLWGKGGKQRLVPVGRPAVEALQAY
Rv2646|M.tuberculosis_H37Rv GEAGLRRAEAAQAHTGDLMDGG-LLLVHGKGGKRRIVPIS-----DYLAA
TH_2517|M.thermoresistible__bu TFEGLRTFEIAKVRGEHFDLVSRTMTVTGKGGITATLPLHHRVVEHAYRM
* * * . .. . : : **** :*:
MMAR_2506|M.marinum_M LVRGRPELARRGRG---TPAIFLNARGGRLSRQSAWQVLQDAAERAGITS
MUL_1690|M.ulcerans_Agy99 LVRGRPELARRGRG---TPAIFLNARGGRLSRQSAWQVLQDAAERADITS
Mb1727|M.bovis_AF2122/97 LVRGRPDLARRGRG---TAAIFLNARGGRLSRQSAWQVLQDAAERAGITA
MLBr_01365|M.leprae_Br4923 LVRGRSDLARRGPGMLATPAIFLNARGGRLSRQSAWQVLQDAAEHAGITS
MAV_3070|M.avium_104 LVRGRSELARRGRG---TPAIFLNVRGGRLSRQSAWQVLQDAAERAGITS
Mflv_3500|M.gilvum_PYR-GCK LVRGRPDLARRGRG---TPAIFLNARGGRLSRQSAWQVLQDAAGRAGVTA
Mvan_3281|M.vanbaalenii_PYR-1 LVRGRPDLARRGRG---TPAIFLNARGGRLSRQSAWQVLQDAAERAGITA
MSMEG_3744|M.smegmatis_MC2_155 LVRGRPDLARRGRG---TPAIFLNARGGRLSRQSAWQVLQDSAERAGIAS
MAB_2366|M.abscessus_ATCC_1997 LVRGRPDLARRGRG--GVPALFLNSRGGRLSRQSAWQVLADAAERAKISA
Rv2646|M.tuberculosis_H37Rv LIRDTP------HG-----YLFPNGTGGHLTAEHVGKLVSRALP-----G
TH_2517|M.thermoresistible__bu PR----------KG-----YWFPGRNGGHQRRESVGQTIKEAITRAG--A
* * . **: : . : : : .
MMAR_2506|M.marinum_M GVSPHMLRHSFATHLLEGGADVRVVQELLGHASVTTTQIYTLVTVH----
MUL_1690|M.ulcerans_Agy99 GVSPHMLRHSFATHLLEGGADVRVVQELLGHASVTTTQIYTLVTVH----
Mb1727|M.bovis_AF2122/97 GVSPHMLRHSFATHLLEGGADVRVVQELLGHASVTTTQIYTLVTVH----
MLBr_01365|M.leprae_Br4923 GVSPHMLRHSFATHLLEGGADIRVVQELMGHASVTTTQIYTLVTVQ----
MAV_3070|M.avium_104 GVSPHMLRHSFATHLLEGGADVRVVQELLGHASVTTTQIYTMVTVH----
Mflv_3500|M.gilvum_PYR-GCK TVSPHVLRHSFATHLLDGGADVRVVQELLGHASVTTTQIYTMVTVN----
Mvan_3281|M.vanbaalenii_PYR-1 TVSPHVLRHSFATHLLDGGADVRVVQELLGHASVTTTQIYTMVTVT----
MSMEG_3744|M.smegmatis_MC2_155 AVSPHTLRHSFATHLLDGGADVRVVQELLGHASVTTTQIYTMVTVH----
MAB_2366|M.abscessus_ATCC_1997 AVSPHTLRHSFATHLLEGGADVRVVQELLGHASVTTTQIYTLVTVS----
Rv2646|M.tuberculosis_H37Rv DATMHTLRHRYATRAYRGSHNLRAVQQLLGHASIVTTERYTALCDD----
TH_2517|M.thermoresistible__bu VGSAHQLRHWFGTALVEAGVDLRTVQTLLRHQNLSSTEIYTQVKEPRRAD
: * *** :.* .. ::*.** *: * .: :*: ** :
MMAR_2506|M.marinum_M ----------ALREVWAGAHPRAT
MUL_1690|M.ulcerans_Agy99 ----------ALREVWAGAHPRAT
Mb1727|M.bovis_AF2122/97 ----------ALREVWAGAHPRAR
MLBr_01365|M.leprae_Br4923 ----------ALREVWAGAHPRAK
MAV_3070|M.avium_104 ----------ALREVWAEAHPRAR
Mflv_3500|M.gilvum_PYR-GCK ----------ALREVWAGAHPRAR
Mvan_3281|M.vanbaalenii_PYR-1 ----------ALREVWAGAHPRAR
MSMEG_3744|M.smegmatis_MC2_155 ----------TLREVWAGAHPRAQ
MAB_2366|M.abscessus_ATCC_1997 ----------ALREVWAGAHPRAR
Rv2646|M.tuberculosis_H37Rv ----------EVRAAAAAAW----
TH_2517|M.thermoresistible__bu AIQLLDPFQLDMVALSASPWKEAA
: * .