For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GVGAVLDGQLQGYLDHLAIERGVAANTLSSYRRDLRRYAEHLKARGVGDLAAVTETDVSEFLVALRLGDP DGGVVPLSAVSAARALIAVRGLHRFAAAEGIAAIDVARDVKPPTPSRRLPKSLSIDEVLALLEAAGGDSE ADSPLTLRNRALLELLYSTGARISEAVGLDIDDVDTSARSVLLRGKGGKQRLIPIGRPAVTALETYLVRG RPDLARRGRGTPAIFLNARGGRLSRQSAWQVLQDAAGRAGVTATVSPHVLRHSFATHLLDGGADVRVVQE LLGHASVTTTQIYTMVTVNALREVWAGAHPRAR*
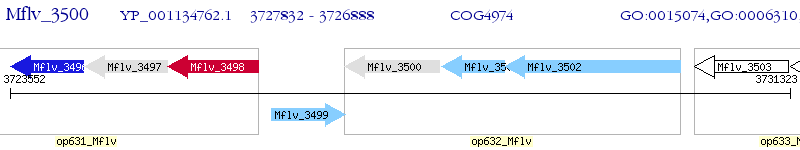
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3500 | xerD | - | 100% (314) | site-specific tyrosine recombinase XerD |
| M. gilvum PYR-GCK | Mflv_4135 | xerC | 1e-54 | 41.56% (308) | site-specific tyrosine recombinase XerC |
| M. gilvum PYR-GCK | Mflv_4645 | - | 2e-22 | 29.13% (333) | phage integrase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1727 | xerD | 1e-147 | 85.57% (305) | site-specific tyrosine recombinase XerD |
| M. tuberculosis H37Rv | Rv1701 | xerD | 1e-147 | 85.57% (305) | site-specific tyrosine recombinase XerD |
| M. leprae Br4923 | MLBr_01365 | xerD | 1e-139 | 79.55% (313) | site-specific tyrosine recombinase XerD |
| M. abscessus ATCC 19977 | MAB_2366 | xerD | 1e-134 | 78.71% (310) | site-specific tyrosine recombinase XerD |
| M. marinum M | MMAR_2506 | xerD | 1e-148 | 85.34% (307) | integrase/recombinase, XerD |
| M. avium 104 | MAV_3070 | xerD | 1e-146 | 84.74% (308) | site-specific tyrosine recombinase XerD |
| M. smegmatis MC2 155 | MSMEG_3744 | xerD | 1e-151 | 86.77% (310) | site-specific tyrosine recombinase XerD |
| M. thermoresistible (build 8) | TH_1542 | xerD | 1e-151 | 87.06% (309) | tyrosine recombinase XerD |
| M. ulcerans Agy99 | MUL_1690 | xerD | 1e-147 | 85.02% (307) | site-specific tyrosine recombinase XerD |
| M. vanbaalenii PYR-1 | Mvan_3281 | xerD | 1e-157 | 90.97% (310) | site-specific tyrosine recombinase XerD |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1727|M.bovis_AF2122/97 ------MKT--------LALQ--LQGYLDHLTIERGVAANTLSSYRRDLR
Rv1701|M.tuberculosis_H37Rv ------MKT--------LALQ--LQGYLDHLTIERGVAANTLSSYRRDLR
MMAR_2506|M.marinum_M ------MST--------LTLDTQLQGYLDHLAIERGVAANTLSSYRRDLR
MUL_1690|M.ulcerans_Agy99 ------MST--------LTLDTQLQGYLDHLAIERGVAANTLSSYRRDLR
MLBr_01365|M.leprae_Br4923 ------MTT--------AALQTQLQGYLDYLIIERSIAANTLSSYRRDLI
MAV_3070|M.avium_104 ------MTT--------VALETQLQGYLDHLTIERGVAANTLSSYRRDLR
Mflv_3500|M.gilvum_PYR-GCK ----MGVG---------AVLDGQLQGYLDHLAIERGVAANTLSSYRRDLR
Mvan_3281|M.vanbaalenii_PYR-1 -MT-TGVDP--------AALDDQFQGYLDHLAIERGVAANTLSSYRRDLR
MSMEG_3744|M.smegmatis_MC2_155 -MTGTSALR--------AVLDEQVQGYLDHLTIERGVAANTLSSYRRDLR
TH_1542|M.thermoresistible__bu VTTSEQVPRSDEPPHPHRVLDDQIQGYLDHLSIERGVAANTLSSYRRDLR
MAB_2366|M.abscessus_ATCC_1997 ----MTAAVG--------LLDGEIQSYLDHLDVERGVAANTLSSYRRDLR
*: .*.***:* :**.:************
Mb1727|M.bovis_AF2122/97 RYSKHLEERGITDLAKVGEHDVSEFLVALRRGDPDSGTAALSAVSAARAL
Rv1701|M.tuberculosis_H37Rv RYSKHLEERGITDLAKVGEHDVSEFLVALRRGDPDSGTAALSAVSAARAL
MMAR_2506|M.marinum_M RYSKHLEDRGITDLAKVGEDDVSEFLVALRRGDPESGVLGLSAVSAARAL
MUL_1690|M.ulcerans_Agy99 RYSKHLEDRGITDLAKVGEDDVSEFLVALRRGDPESGVLGLSAVSAARAL
MLBr_01365|M.leprae_Br4923 RYSKHLSDRGIEDLAKVGEHDVSEFLVALRRGDPDSGVAALSAVSAARAL
MAV_3070|M.avium_104 RYTKHLSDRGISDLAKVGEDDVSDFLVALRRGDPDTGAAALSAVSAARAL
Mflv_3500|M.gilvum_PYR-GCK RYAEHLKARGVGDLAAVTETDVSEFLVALRLGDPDGGVVPLSAVSAARAL
Mvan_3281|M.vanbaalenii_PYR-1 RYAEHLASRGVTDLAGVAEADVSEFLVSLRRGDPDNGVVPLSAVSAARAV
MSMEG_3744|M.smegmatis_MC2_155 RYAEHLRQRGIDDLARVAESDVSDFLVALRRGDPDAGVTALSAVSAARAV
TH_1542|M.thermoresistible__bu RYAEHLLARGIDDLAKVHESDVADFLVALRRGDPDTGAAGLSAVSAARAL
MAB_2366|M.abscessus_ATCC_1997 RYQQHLADRKIDRLAEVCETDVSDFVVTLRRGDPANGVPELSASSAARAL
** :** * : ** * * **::*:*:** *** *. *** *****:
Mb1727|M.bovis_AF2122/97 IAVRGLHRFAAAEGLAELDVARAVRPPTPSRRLPKSLTIDEVLSLLEGAG
Rv1701|M.tuberculosis_H37Rv IAVRGLHRFAAAEGLAELDVARAVRPPTPSRRLPKSLTIDEVLSLLEGAG
MMAR_2506|M.marinum_M IAVRGLHRFAAAEGLAALDVARAVRPPTPGRRLPKSLTIDEVLALLEGAG
MUL_1690|M.ulcerans_Agy99 IAVRGLHRFAAAEGLAALDVARAVRPPTPGRRLPKSLTIDEVLALLEGAG
MLBr_01365|M.leprae_Br4923 IAVRGLHRFAVAEGLVDLDVARAVRPPTPGRRLPKSLTVDEVLALLESVG
MAV_3070|M.avium_104 IAVRGLHRFLAAEGLAELDVARAVRPPTPGRRLPKSLTIDQVLALLEAAG
Mflv_3500|M.gilvum_PYR-GCK IAVRGLHRFAAAEGIAAIDVARDVKPPTPSRRLPKSLSIDEVLALLEAAG
Mvan_3281|M.vanbaalenii_PYR-1 IAVRGLHRFAAAEGITTVDVAREVKPPTPGRRLPKSLSIDEVLALLEAAG
MSMEG_3744|M.smegmatis_MC2_155 VAVRGLHRFAAAEGLTENDVARAVKPPTPSRRLPKSLTLDEVLALLEAAG
TH_1542|M.thermoresistible__bu IAVRGLHRFAAAEGVTELDVARSVKPPTPSRRLPKSLTVDEVLALLAGAG
MAB_2366|M.abscessus_ATCC_1997 IAVRGLHRFAAIEGLAPTDVARAVKPPTPNRRLPKSLTVEQVEALLNAAG
:******** . **:. **** *:****.*******::::* :** ..*
Mb1727|M.bovis_AF2122/97 GDKPSDGPLTLRNRAVLELLYSTGARISEAVGLDLDDIDTHARSVLLRGK
Rv1701|M.tuberculosis_H37Rv GDKPSDGPLTLRNRAVLELLYSTGARISEAVGLDLDDIDTHARSVLLRGK
MMAR_2506|M.marinum_M GDNPADGPLTLRNRALLELLYSTGSRISEAVGLDVDDIDTQARTVLLQGK
MUL_1690|M.ulcerans_Agy99 GDNPADGPLTLRNRALLELLYSTGSRISEAVGLDVDDIDTQARTVLLQGK
MLBr_01365|M.leprae_Br4923 GESRADGPLVLRNRALLELLYSTGSRISEAVGLDVDDVDTQARTVLLQGK
MAV_3070|M.avium_104 GESPADGPLTLRNRALLELLYSTGARISEAVGLDVDDVDTQARSVLLRGK
Mflv_3500|M.gilvum_PYR-GCK GDSEADSPLTLRNRALLELLYSTGARISEAVGLDIDDVDTSARSVLLRGK
Mvan_3281|M.vanbaalenii_PYR-1 GDGEADGPLTLRNRALLELLYSTGARISEAVGLDVDDVDTEARSVLLRGK
MSMEG_3744|M.smegmatis_MC2_155 GDSEADSPLTLRNRALLELLYSTGARISEAVGLDVDDVDTQARSVLLHGK
TH_1542|M.thermoresistible__bu GDSESDGPLTLRNRALLELLYSTGARISEAVGLDIDDVDTVARSVLLRGK
MAB_2366|M.abscessus_ATCC_1997 GGG-SDGPLDLRNRALLELLYSTGARISEAVGLDVDDVDVQARSVLLWGK
* :*.** *****:********:*********:**:*. **:*** **
Mb1727|M.bovis_AF2122/97 GGKQRLVPVGRPAVHALDAYLVRGRPDLARRGRG---TAAIFLNARGGRL
Rv1701|M.tuberculosis_H37Rv GGKQRLVPVGRPAVHALDAYLVRGRPDLARRGRG---TAAIFLNARGGRL
MMAR_2506|M.marinum_M GGKQRLVPVGRPAVQALDAYLVRGRPELARRGRG---TPAIFLNARGGRL
MUL_1690|M.ulcerans_Agy99 GGKQRLVPVGRPAVQALDAYLVRGRPELARRGRG---TPAIFLNARGGRL
MLBr_01365|M.leprae_Br4923 GGKQRLVPVGRPAVQALDAYLVRGRSDLARRGPGMLATPAIFLNARGGRL
MAV_3070|M.avium_104 GGKQRLVPIGRPAVAALDAYLVRGRSELARRGRG---TPAIFLNVRGGRL
Mflv_3500|M.gilvum_PYR-GCK GGKQRLIPIGRPAVTALETYLVRGRPDLARRGRG---TPAIFLNARGGRL
Mvan_3281|M.vanbaalenii_PYR-1 GGKQRLIPIGRPAVSALDNYLVRGRPDLARRGRG---TPAIFLNARGGRL
MSMEG_3744|M.smegmatis_MC2_155 GGKQRLVPIGRPAVSALDAYLVRGRPDLARRGRG---TPAIFLNARGGRL
TH_1542|M.thermoresistible__bu GGKQRLVPIGRPAVSALDAYLVRGRPELVRRGRG---TPAIFLNARGGRL
MAB_2366|M.abscessus_ATCC_1997 GGKQRLVPVGRPAVEALQAYLVRGRPDLARRGRG--GVPALFLNSRGGRL
******:*:***** **: ******.:*.*** * ..*:*** *****
Mb1727|M.bovis_AF2122/97 SRQSAWQVLQDAAERAGITAGVSPHMLRHSFATHLLEGGADVRVVQELLG
Rv1701|M.tuberculosis_H37Rv SRQSAWQVLQDAAERAGITAGVSPHMLRHSFATHLLEGGADVRVVQELLG
MMAR_2506|M.marinum_M SRQSAWQVLQDAAERAGITSGVSPHMLRHSFATHLLEGGADVRVVQELLG
MUL_1690|M.ulcerans_Agy99 SRQSAWQVLQDAAERADITSGVSPHMLRHSFATHLLEGGADVRVVQELLG
MLBr_01365|M.leprae_Br4923 SRQSAWQVLQDAAEHAGITSGVSPHMLRHSFATHLLEGGADIRVVQELMG
MAV_3070|M.avium_104 SRQSAWQVLQDAAERAGITSGVSPHMLRHSFATHLLEGGADVRVVQELLG
Mflv_3500|M.gilvum_PYR-GCK SRQSAWQVLQDAAGRAGVTATVSPHVLRHSFATHLLDGGADVRVVQELLG
Mvan_3281|M.vanbaalenii_PYR-1 SRQSAWQVLQDAAERAGITATVSPHVLRHSFATHLLDGGADVRVVQELLG
MSMEG_3744|M.smegmatis_MC2_155 SRQSAWQVLQDSAERAGIASAVSPHTLRHSFATHLLDGGADVRVVQELLG
TH_1542|M.thermoresistible__bu SRQSAWQVLQDAAERAGIKAGVSPHTLRHSFATHLLDGGADVRVVQELLG
MAB_2366|M.abscessus_ATCC_1997 SRQSAWQVLADAAERAKISAAVSPHTLRHSFATHLLEGGADVRVVQELLG
********* *:* :* : : **** **********:****:******:*
Mb1727|M.bovis_AF2122/97 HASVTTTQIYTLVTVHALREVWAGAHPRAR
Rv1701|M.tuberculosis_H37Rv HASVTTTQIYTLVTVHALREVWAGAHPRAR
MMAR_2506|M.marinum_M HASVTTTQIYTLVTVHALREVWAGAHPRAT
MUL_1690|M.ulcerans_Agy99 HASVTTTQIYTLVTVHALREVWAGAHPRAT
MLBr_01365|M.leprae_Br4923 HASVTTTQIYTLVTVQALREVWAGAHPRAK
MAV_3070|M.avium_104 HASVTTTQIYTMVTVHALREVWAEAHPRAR
Mflv_3500|M.gilvum_PYR-GCK HASVTTTQIYTMVTVNALREVWAGAHPRAR
Mvan_3281|M.vanbaalenii_PYR-1 HASVTTTQIYTMVTVTALREVWAGAHPRAR
MSMEG_3744|M.smegmatis_MC2_155 HASVTTTQIYTMVTVHTLREVWAGAHPRAQ
TH_1542|M.thermoresistible__bu HASVTTTQIYTMVTVHALREVWAGAHPRAR
MAB_2366|M.abscessus_ATCC_1997 HASVTTTQIYTLVTVSALREVWAGAHPRAR
***********:*** :****** *****