For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
REHLMSIEVANESGIDVSEAELVSVARFVIAKMDVNPCAELSMLLLDTAAMADLHMRWMDLPGPTDVMSF PMDELEPGGRPDAPEPGPSMLGDIVLCPEFAAEQAAAAGHSLGHELALLTIHGVLHLLGYDHAEPDEEKE MFALQDRLLEEWVADQVEAYQHDRQDEKDRRLLDKSRYFDL*
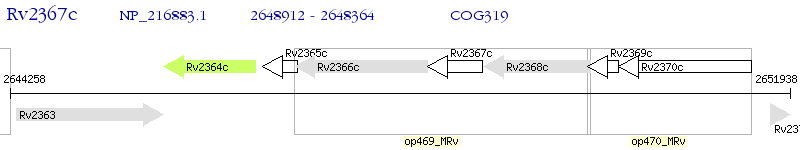
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2367c | - | - | 100% (182) | hypothetical protein Rv2367c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2388c | - | 1e-104 | 100.00% (182) | putative metalloprotease |
| M. gilvum PYR-GCK | Mflv_2707 | - | 3e-90 | 90.34% (176) | putative metalloprotease |
| M. leprae Br4923 | MLBr_00628 | - | 1e-88 | 89.14% (175) | putative metalloprotease |
| M. abscessus ATCC 19977 | MAB_1669 | - | 1e-78 | 79.55% (176) | putative metalloprotease |
| M. marinum M | MMAR_3677 | - | 2e-94 | 93.75% (176) | metal-dependent hydrolase |
| M. avium 104 | MAV_2026 | - | 9e-95 | 93.75% (176) | putative metalloprotease |
| M. smegmatis MC2 155 | MSMEG_4496 | - | 8e-90 | 89.20% (176) | putative metalloprotease |
| M. thermoresistible (build 8) | TH_1055 | - | 9e-88 | 88.07% (176) | UNKNOWN |
| M. ulcerans Agy99 | MUL_3620 | - | 2e-94 | 93.75% (176) | putative metalloprotease |
| M. vanbaalenii PYR-1 | Mvan_3831 | - | 6e-90 | 89.20% (176) | putative metalloprotease |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2707|M.gilvum_PYR-GCK -----MSIEVSNESGIDVSESELVSVARFVIRKMDVNPAAELSMVLLDTA
Mvan_3831|M.vanbaalenii_PYR-1 -----MTIEVSNESGIDVSEEELISVARFVIGKMDVNPAAELSMLLLDTA
Rv2367c|M.tuberculosis_H37Rv MREHLMSIEVANESGIDVSEAELVSVARFVIAKMDVNPCAELSMLLLDTA
Mb2388c|M.bovis_AF2122/97 MREHLMSIEVANESGIDVSEAELVSVARFVIAKMDVNPCAELSMLLLDTA
MLBr_00628|M.leprae_Br4923 -----MSVEVSNESGFDVSEVELVSVARFVIIKMDVNPAAELSMVLLDTA
MMAR_3677|M.marinum_M -----MSIEVSNESGIDVSETELVSVARFVIGKMDVNPGAELSMVLLDTA
MUL_3620|M.ulcerans_Agy99 -----MSIEVSNESGIDVSETELVSVARFVIGKMDVNPGAELSMVLLDTA
MAV_2026|M.avium_104 -----MSIEVSNESGIDVSEAELISVARFVIAKMDVNPAAELSMVLLDTA
MSMEG_4496|M.smegmatis_MC2_155 -----MSIEVSNESGYDVSEPELISVARFVIEKMDVHPAAELSMVLLDSA
TH_1055|M.thermoresistible__bu -----MSIEVSNESGIDVCEEELISVARFVIEKMNVHPAAELSMVLLDPA
MAB_1669|M.abscessus_ATCC_1997 -----MSIEVVNESGIDVAEGELVSVARFAIAAMDVHPAAELSMMLVDLA
*::** **** **.* **:*****.* *:*:* *****:*:* *
Mflv_2707|M.gilvum_PYR-GCK AMADLHMRWMDLPGPTDVMSFPMDELEPGGRPDTPEPGPSMLGDIVLCPE
Mvan_3831|M.vanbaalenii_PYR-1 AMADLHMRWMDLPGPTDVMSFPMDELEPGGRPDAPEPGPSMLGDIVLCPE
Rv2367c|M.tuberculosis_H37Rv AMADLHMRWMDLPGPTDVMSFPMDELEPGGRPDAPEPGPSMLGDIVLCPE
Mb2388c|M.bovis_AF2122/97 AMADLHMRWMDLPGPTDVMSFPMDELEPGGRPDAPEPGPSMLGDIVLCPE
MLBr_00628|M.leprae_Br4923 AMADLHMRWMDLPGPTDVMSFPMDEFEPGGRPDAAEPGPSMLGDIVLCPE
MMAR_3677|M.marinum_M AMADLHMRWMDLPGPTDVMSFPMDELEPGGRPDAPEPGPAMLGDIVLCPE
MUL_3620|M.ulcerans_Agy99 AMADLHMRWMDLPGPTDVMSFPMDELEPGGRPDAPEPGPAMLGDIVLCPE
MAV_2026|M.avium_104 AMADLHMRWMDLPGPTDVMSFPMDELEPGGRPDAPEPGPAMLGDIVLCPE
MSMEG_4496|M.smegmatis_MC2_155 AMADLHMRWMDLPGPTDVMSFPMDELEPGGRPDAPEPGPAMLGDIVLCPE
TH_1055|M.thermoresistible__bu AMADLHMRWMDLPGPTDVMSFPMDELEPGGRPDAAEPGPAMLGDIVLCPE
MAB_1669|M.abscessus_ATCC_1997 AMADLHMRWMDLPGPTDVMSFPMDELEPGGRPDAPEPGPSMLGDIVLCPQ
*************************:*******:.****:*********:
Mflv_2707|M.gilvum_PYR-GCK FAAKQAADAGHTLGQELALLTVHGVLHLLGYDHAEPDEEKEMFALQRELL
Mvan_3831|M.vanbaalenii_PYR-1 FAAKQAADAGHTLGQELALLTVHGVLHLLGYDHAEPDEEKEMFALQRELL
Rv2367c|M.tuberculosis_H37Rv FAAEQAAAAGHSLGHELALLTIHGVLHLLGYDHAEPDEEKEMFALQDRLL
Mb2388c|M.bovis_AF2122/97 FAAEQAAAAGHSLGHELALLTIHGVLHLLGYDHAEPDEEKEMFALQDRLL
MLBr_00628|M.leprae_Br4923 FAAQQAAAEGHSLGHELALLTIHGVLHLLGYDHGEPDEEKEMFALQGRLL
MMAR_3677|M.marinum_M FAAEQAAAAGHSLGHELALLTIHGVLHLLGYDHGEPDEEKEMFALQDRLL
MUL_3620|M.ulcerans_Agy99 FAAEQAAAAGHSLGHELALLTIHGVLHLLGYDHGEPDEEKEMFALQDRLL
MAV_2026|M.avium_104 FAADQAAAAGHSLGHELALLTIHGVLHLLGYDHGEPDEEKEMFALQDRLL
MSMEG_4496|M.smegmatis_MC2_155 FAEQQAAKAGHSLGHELALLTVHGVLHLLGYDHAEPDEEKEMFALQRQLL
TH_1055|M.thermoresistible__bu FAAEQAAAAGHSLGQELALLTVHGVLHLLGYDHAEPEEEKEMFALQRRLL
MAB_1669|M.abscessus_ATCC_1997 FAAEQAEAAGHSLAHELALLTVHGVLHLLGYDHAEPEEEREMFALQNQLL
** .** **:*.:******:***********.**:**:****** .**
Mflv_2707|M.gilvum_PYR-GCK EEWVAEQVEAYHLDRQNEKDRRLLDKSRYF------DTP
Mvan_3831|M.vanbaalenii_PYR-1 EEWVADQVQAYHQDRQSQKDQRLLDKSRYFDELNHGDTP
Rv2367c|M.tuberculosis_H37Rv EEWVADQVEAYQHDRQDEKDRRLLDKSRYFDL-------
Mb2388c|M.bovis_AF2122/97 EEWVADQVEAYQHDRQDEKDRRLLDKSRYFDL-------
MLBr_00628|M.leprae_Br4923 EEWVAEQVRAYQHDRQNEKDCRLLYKSGYFGYL------
MMAR_3677|M.marinum_M EEWVAEQVQAYQQDRQDERDRRLLDKSRYFDEP------
MUL_3620|M.ulcerans_Agy99 EEWVAEQVQAYQQDRQDERDRRLLDKSRYFDEP------
MAV_2026|M.avium_104 EEWVADQVEAYHQDRQQERDRRLLDKSRYFDN-------
MSMEG_4496|M.smegmatis_MC2_155 EEWVADQVEAYHADRQSEKDRRLLDKSRYFDEP------
TH_1055|M.thermoresistible__bu EEWVADQVEAYHQELQSEKDRRLLDKSRYFDQL------
MAB_1669|M.abscessus_ATCC_1997 QDWYEQQAQLY-------RDSRLLDKSRNFDE-------
::* :*.. * :* *** ** *