For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTATVLLEVPFSARGDRIPDAVAELRTREPIRKVRTITGAEAWLVSSYALCTQVLEDRRFSMKETAAAGA PRLNALTVPPEVVNNMGNIADAGLRKAVMKAITPKAPGLEQFLRDTANSLLDNLITEGAPADLRNDFADP LATALHCKVLGIPQEDGPKLFRSLSIAFMSSADPIPAAKINWDRDIEYMAGILENPNITTGLMGELSRLR KDPAYSHVSDELFATIGVTFFGAGVISTGSFLTTALISLIQRPQLRNLLHEKPELIPAGVEELLRINLSF ADGLPRLATADIQVGDVLVRKGELVLVLLEGANFDPEHFPNPGSIELDRPNPTSHLAFGRGQHFCPGSAL GRRHAQIGIEALLKKMPGVDLAVPIDQLVWRTRFQRRIPERLPVLW
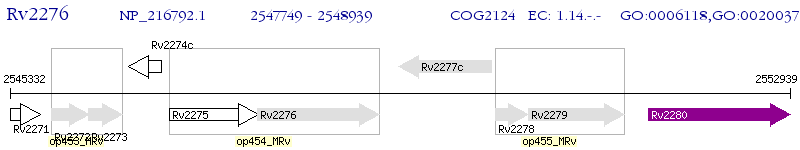
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2276 | cyp121 | - | 100% (396) | CYTOCHROME P450 121 CYP121 |
| M. tuberculosis H37Rv | Rv1880c | cyp140 | 1e-28 | 29.73% (296) | cytochrome p450 140 CYP140 |
| M. tuberculosis H37Rv | Rv3121 | cyp141 | 9e-24 | 23.90% (385) | cytochrome P450 141 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2299 | cyp121 | 0.0 | 100.00% (396) | cytochrome P450 121 CYP121 |
| M. gilvum PYR-GCK | Mflv_0225 | - | 4e-30 | 27.30% (370) | cytochrome P450 |
| M. leprae Br4923 | MLBr_02088 | - | 1e-30 | 32.28% (316) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_4457 | - | 8e-29 | 26.59% (361) | putative cytochrome P450 |
| M. marinum M | MMAR_2768 | cyp140A5 | 2e-28 | 29.05% (296) | cytochrome P450 140A5 Cyp140A5 |
| M. avium 104 | MAV_2156 | - | 2e-32 | 27.55% (392) | P450 heme-thiolate protein |
| M. smegmatis MC2 155 | MSMEG_0762 | - | 5e-35 | 27.84% (370) | cytochrome P450 FAS1 |
| M. thermoresistible (build 8) | TH_2999 | - | 2e-29 | 28.46% (369) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_2979 | cyp140A5 | 2e-28 | 29.05% (296) | cytochrome P450 140A5 Cyp140A5 |
| M. vanbaalenii PYR-1 | Mvan_0600 | - | 9e-30 | 27.51% (378) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4457|M.abscessus_ATCC_1997 ----MQHRIKQRTHWAVTHGIGRAYLKVLARRG-EPVAQLGIDVGQAPDI
MAV_2156|M.avium_104 ------MRVRLGARWMAMHGLPRAYFAVQARRG-DPLARLLRSGTTGEDR
MMAR_2768|M.marinum_M --------MKDKLHWFAMHGVIRGMANIGLRRG-ELEARLIADPAVAANP
MUL_2979|M.ulcerans_Agy99 --------MKDKLHWFAMHGVIRGMANIGLRRG-ELEARLIADPAVAANP
TH_2999|M.thermoresistible__bu ---------------LSLHGVIRALTAYGARRG-DPQARLIADPAVRADP
Mvan_0600|M.vanbaalenii_PYR-1 ----MAADLQQRIRWLALHGVIRGLSKVAVRRGQDPQARLIADPAVRADP
Mflv_0225|M.gilvum_PYR-GCK --------------------------MSQATGEAAPDLPPLHMRRDGFDP
MSMEG_0762|M.smegmatis_MC2_155 --------------------------MTQAQ-----ALPPLHIRRDAFDP
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
Rv2276|M.tuberculosis_H37Rv -----------------------MTATVLLEVPFSARGDRIPDAVAELRT
Mb2299|M.bovis_AF2122/97 -----------------------MTATVLLEVPFSARGDRIPDAVAELRT
MAB_4457|M.abscessus_ATCC_1997 YRIIDKIRERGRLSRAGDGWITADAQIVRTIFRDNRFVTFKPEHRSAS-P
MAV_2156|M.avium_104 YALMEQIRARGPLMRAPFVWASVDHAVCRQVLRDKRFGVTSPTEMELP-R
MMAR_2768|M.marinum_M GPFHDELRRHGRMVRTRVNYLTVDYQLAHDVLRSDDFHTIS-FGENLP-G
MUL_2979|M.ulcerans_Agy99 GPFHDELRRHGRMVRTRVNYLTVDYQLAHDVLRSDDFHTIS-FGENLP-G
TH_2999|M.thermoresistible__bu AAFADELRGRGPLIRGRAALLTFDHGVANELLRSDDFRVSE-LGGNLP-A
Mvan_0600|M.vanbaalenii_PYR-1 AAFADELRGRGPVIRCRAVLMTVDHQVVNEILRSDDFRVSS-LGAGLP-K
Mflv_0225|M.gilvum_PYR-GCK TPHLREIREREGVRSVTNAFGMQVYLFTRYDDIKTVLSDHSRFSNGRPPG
MSMEG_0762|M.smegmatis_MC2_155 TPELGEIRAGEGVHVTVNPFGMQVYLVTRHEDVKTVLSDHERFSNSRPPG
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQLPGMPLTVFSSFSDCDEALRHPLSASDRLKATLAQQ
Rv2276|M.tuberculosis_H37Rv REPIRKVR----TITGAEAWLVSSYALCTQVLEDRRFSMKETAAAGAP--
Mb2299|M.bovis_AF2122/97 REPIRKVR----TITGAEAWLVSSYALCTQVLEDRRFSMKETAAAGAP--
: : . .
MAB_4457|M.abscessus_ATCC_1997 IIQRLAAWSDPQLLNPAEPPSILITDPPDHGRLRRLVAAPFTPRAIEGLR
MAV_2156|M.avium_104 PVRALIARTDPGVANPVAPPAMVIVDPPDHTRYRQLVAQSFTPRAIEALN
MMAR_2768|M.marinum_M PLRWLERRTRDEQLHPLRPPSLLAVEPPDHTRYRKTVSAVFTSRAVAALR
MUL_2979|M.ulcerans_Agy99 PLRWLERRTRDEQLHPLRPPSLLAVEPPDHTRYRKTVSAVFTSRAVAALR
TH_2999|M.thermoresistible__bu PLRWVAAKADTGHLHPIQPPSLLSIEPPDHTRLRKLVSSVFTTRAVATLK
Mvan_0600|M.vanbaalenii_PYR-1 PLQWINRKTHPGLLHPIEPPSLLSIEPPDHTRCRKLVSSVFTTRAVAALR
Mflv_0225|M.gilvum_PYR-GCK FVVPGAPPVDEETQARARAGNLLGLDPPEHQRLRRMLTPEFTHRRMQRLR
MSMEG_0762|M.smegmatis_MC2_155 FVLPGAPQISAEEQASNRAGNLLGLDPPEHQRLRRMLTPEFTIRRIKRLE
MLBr_02088|M.leprae_Br4923 AIAAGAEPR------PFYASSFMFLDPPDHTRLRKLVSKAFAPKVVQALE
Rv2276|M.tuberculosis_H37Rv ----------RLNALTVPPEVVNNMGNIADAGLRKAVMKAITPKAPG-LE
Mb2299|M.bovis_AF2122/97 ----------RLNALTVPPEVVNNMGNIADAGLRKAVMKAITPKAPG-LE
. . . *: : :: : *.
MAB_4457|M.abscessus_ATCC_1997 DRIQEVTNGHLDVLQQR---QSPDLIADFTAKIPIAVIGEMIAVPPPDYS
MAV_2156|M.avium_104 TRVAQVTMELIERIAAT---PQPDLIADFATRLPVAIIAEILGMPPDSYP
MMAR_2768|M.marinum_M DRVEQTANSLLDQLAQR--PGVVDVVGQYCSQLPIMVISEILGVPEQDRG
MUL_2979|M.ulcerans_Agy99 DRVEQTANSLLDQLAQR--PGVVDVVGQYCSQLPIMVISEILGVPEQDRG
TH_2999|M.thermoresistible__bu ERLQQTAFDLLDGLDEVGASDVVDVIDRYCSQLPIAVISDILGVPDEDRD
Mvan_0600|M.vanbaalenii_PYR-1 ERVQQTANDLLDDLERE--SGVVDIVERYCSQLPVAVISDILGVPERDRR
Mflv_0225|M.gilvum_PYR-GCK PRIAEIVDTQIDALAAAGR--PADLVEHFALPIPSLVICELLGVPYADRD
MSMEG_0762|M.smegmatis_MC2_155 PRIVEIVDAHLDAMESAGP--PADIVADFALPIPSLVICELLGVPYEDRT
MLBr_02088|M.leprae_Br4923 GDIAALVDSLLDKGAAAG---QFDVIADLAFPLAVAVICRLLGVPYEDAP
Rv2276|M.tuberculosis_H37Rv QFLRDTANSLLDNLITEG--APADLRNDFADPLATALHCKVLGIPQEDGP
Mb2299|M.bovis_AF2122/97 QFLRDTANSLLDNLITEG--APADLRNDFADPLATALHCKVLGIPQEDGP
: . :: *: :. : ::.:* .
MAB_4457|M.abscessus_ATCC_1997 RLYTAMNRAIQLIATTA----PSWSEYQDGTAALREIDEYLEKHVARLRR
MAV_2156|M.avium_104 RML-AWGRSGSPLLDLG----IDWKTYRDAIAGLRGVDEYLLAHFHQLRA
MMAR_2768|M.marinum_M RVL-EYGELAAPSLDIG----VPYRQYRRIQQGITGFNSWLTAHLEQLRR
MUL_2979|M.ulcerans_Agy99 RVL-EYGELAAPSLDIG----VTYRQYRRIQQGITGFNSWLTAHLEQLRR
TH_2999|M.thermoresistible__bu RIL-HFGELGAPSLDIG----LSWRQYRQVREGIVGFNTWLTAHLQRLRR
Mvan_0600|M.vanbaalenii_PYR-1 HIL-HFGELGAPSLDIG----LSWRQYTQVHEGLVGFNEWLGGHMDELRR
Mflv_0225|M.gilvum_PYR-GCK DFQRRSARQLDLSIPIP-----------ERLELQREGREYMAALVAGARR
MSMEG_0762|M.smegmatis_MC2_155 DFQQRSARQLDLSAPMP-----------ERLELQRQGRAYMRGLVERSRT
MLBr_02088|M.leprae_Br4923 EFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVKCRRG
Rv2276|M.tuberculosis_H37Rv KLFRSLSIAFMSSADPIP----------AAKINWDRDIEYMAGILE--NP
Mb2299|M.bovis_AF2122/97 KLFRSLSIAFMSSADPIP----------AAKINWDRDIEYMAGILE--NP
. :: . .
MAB_4457|M.abscessus_ATCC_1997 E-GTESE--LATALLDSD----LSHFELKMFFAVFLGAGFVTTTHLMGKA
MAV_2156|M.avium_104 DPHSDNP--FGRMAADGS----LTDRELTANAALIVGAGFETTVNLIGNG
MMAR_2768|M.marinum_M NPGDDLMSQLIRVAESGDAGTYLDESELRAVAGLVLVAGFETTVNLLGNG
MUL_2979|M.ulcerans_Agy99 NPGDDLMSQLIRVAESGDAGTYLDKSELRAIAGLVLVAGFETTVNLLGNG
TH_2999|M.thermoresistible__bu DPGEDLLSQLIQAADAET-----TERELEALAGLVLAAGFETTVNLLGNG
Mvan_0600|M.vanbaalenii_PYR-1 NPGDDLMSQLIQASQAADEGSRLTDRELQATAGLVLAAGFETTVNLLGNG
Mflv_0225|M.gilvum_PYR-GCK HPGDDILG--MLIREHGGE---LTDDELIGVASLLLLAGHETTSNMLALG
MSMEG_0762|M.smegmatis_MC2_155 RPGDDILG--MLVREHGTE---LTDDELIGIAGLLLLAGHETTSNMLGLG
MLBr_02088|M.leprae_Br4923 TPGEDLISRLIELDESGDQ---LTEEEIIATCGLLLVAGHETTVNLIANA
Rv2276|M.tuberculosis_H37Rv NITTGLMGELSRLRKDPAYS-HVSDELFATIGVTFFGAGVISTGSFLTTA
Mb2299|M.bovis_AF2122/97 NITTGLMGELSRLRKDPAYS-HVSDELFATIGVTFFGAGVISTGSFLTTA
. : .. ** :* :: .
MAB_4457|M.abscessus_ATCC_1997 IVTLLRHPEQLALLQADPSLWPNAVEELMRYDTSNQ-WSARVATETVEIE
MAV_2156|M.avium_104 IVLLLRHPEQLALLHDNPDLWPSAVEEILRIASPVQ-MTARTPACDVDIA
MMAR_2768|M.marinum_M IRMLLDAPEQLETLRQRPELWPNAVEEILRLDSPVQ-LTARVARTDVEIA
MUL_2979|M.ulcerans_Agy99 IRMLLDAPEQLETLRQRPELWPNAVEEILRLDSPVQ-LTARVARTDVEIA
TH_2999|M.thermoresistible__bu IRMLLDTPGHLDTLAARPELWPNAVEEILRLDPPVQ-MTARVARVDTDIA
Mvan_0600|M.vanbaalenii_PYR-1 IRMLLETPGHLETLAARPELWPNAVEEILRLDSPVQ-MSARFARRDVTVG
Mflv_0225|M.gilvum_PYR-GCK VLALLRHPDQLAAIRDDPAAVAPAVEELLRWLSIVQTSIPRITTTDVEIA
MSMEG_0762|M.smegmatis_MC2_155 VLALLRHPDQLACVRDDPDAVGPAIEELLRWLSIVSTALPRITTTDVELA
MLBr_02088|M.leprae_Br4923 VLAMLRNPSQWKALSSNPQRAPLVVEETLRYDPAIH-LIGRVAAKDMTIG
Rv2276|M.tuberculosis_H37Rv LISLIQRPQLRNLLHEKPELIPAGVEELLRINLSFADGLPRLATADIQVG
Mb2299|M.bovis_AF2122/97 LISLIQRPQLRNLLHEKPELIPAGVEELLRINLSFADGLPRLATADIQVG
: :: * : * :** :* * . :
MAB_4457|M.abscessus_ATCC_1997 GHTIEAGQSALLLLGGANRDPAAFENPDVFDITRPNARENITLGTGIHVC
MAV_2156|M.avium_104 GAHIGAGEMVGLFLGGANRDPKVFSDPTTFDVTRPNAREHLAFASGIHAC
MMAR_2768|M.marinum_M SWPINRGEMVVAYLAGANRDPAVFPNPHRFDVERANAGRHLAFSTGRHFC
MUL_2979|M.ulcerans_Agy99 SWPINRGEMVVAYLAGANRDPAVFPNPHRFDVERANAGRHLAFSTGRHFC
TH_2999|M.thermoresistible__bu GTAVRRGEMVIIHLAGANRDPKVFADPHRFDIERANAHRHLSFSGGRHFC
Mvan_0600|M.vanbaalenii_PYR-1 GTEIKRGELVILHLAGANRDPEVFTDPHRFDIERDNAGRHLSFSGGRHFC
Mflv_0225|M.gilvum_PYR-GCK GVAIPAGQLVFASLPAGNRDPEFIEHPEVLDIGRG-APGHLAFGHGVHHC
MSMEG_0762|M.smegmatis_MC2_155 GVTIPAGHLVFASLPAGNRDPEFIDDPDTLDIRRG-APGHLAFGHGVHHC
MLBr_02088|M.leprae_Br4923 QTTLTEGDTMVLLLAAANRDPAVYSRPDEFDPDRP-SSRHLAFAVGSHFC
Rv2276|M.tuberculosis_H37Rv DVLVRKGELVLVLLEGANFDPEHFPNPGSIELDRPNPTSHLAFGRGQHFC
Mb2299|M.bovis_AF2122/97 DVLVRKGELVLVLLEGANFDPEHFPNPGSIELDRPNPTSHLAFGRGQHFC
: *. * ..* ** * :: * . ::::. * * *
MAB_4457|M.abscessus_ATCC_1997 LGQVLARAELHTALQTLFERFPRLSLAG---EPEYLNGMGIHGLRLLPVT
MAV_2156|M.avium_104 LGAALARIEGATALRALFENFPDLRLTA---APQRRSLINLHGYTRLPAQ
MMAR_2768|M.marinum_M LGAALARAEGEVGLRTFFERFPDVRAAG---AGSRRDTRVLRGWSTLPVT
MUL_2979|M.ulcerans_Agy99 LGAALARAEGEVGLRTFFERFPDVRAAG---AGSRRDTRVLRGWSTLPVT
TH_2999|M.thermoresistible__bu LGAALARAEGEVGLRTFFQRYPKARSAG---GGDRRDTRVLRGWASLPIT
Mvan_0600|M.vanbaalenii_PYR-1 LGAALARAEGEVGLRTFFERYPDARLAG---QGSRRDTRVLRGWSTLPIT
Mflv_0225|M.gilvum_PYR-GCK LGAPLARMEMAIAFPALFRRFPSLEPAEDFADVAFRSFHFIYGLKSLEVS
MSMEG_0762|M.smegmatis_MC2_155 LGAPLARMEMRIALPALLRRFPTLALAEPFEDVRWRPFHFIYGLQSLAVA
MLBr_02088|M.leprae_Br4923 LGAALARLEATVTLSAISARFPQVQLAG---ELVYKPNVAMRGMSALPVQ
Rv2276|M.tuberculosis_H37Rv PGSALGRRHAQIGIEALLKKMPGVDLAVPIDQLVWRTRFQRRIPERLPVL
Mb2299|M.bovis_AF2122/97 PGSALGRRHAQIGIEALLKKMPGVDLAVPIDQLVWRTRFQRRIPERLPVL
* *.* . : :: . * : *
MAB_4457|M.abscessus_ATCC_1997 LG-----------
MAV_2156|M.avium_104 LGGRRTTSTTIPV
MMAR_2768|M.marinum_M LGPARALAAS---
MUL_2979|M.ulcerans_Agy99 LGPARALAAS---
TH_2999|M.thermoresistible__bu LGPARTTVGS---
Mvan_0600|M.vanbaalenii_PYR-1 LGTARAALGS---
Mflv_0225|M.gilvum_PYR-GCK W------------
MSMEG_0762|M.smegmatis_MC2_155 W------------
MLBr_02088|M.leprae_Br4923 V------------
Rv2276|M.tuberculosis_H37Rv W------------
Mb2299|M.bovis_AF2122/97 W------------