For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RWGVESICTQLTELGVPIAPSTYYDHINREPSRRELRDGELKEHISRVHAANYGVYGARKVWLTLNREGI EVARCTVERLMTKLGLSGTTRGKARRTTIADPATARPADLVQRRFGPPAPNRLWVADLTYVSTWAGFAYV AFVTDAYARRILGWRVASTMATSMVLDAIEQAIWTRQQEGVLDLKDVIHHTDRGSQYTSIRFSERLAEAG IQPSVGAVGSSYDNALAETINGLYKTELIKPGKPWRSIEDVELATARWVDWFNHRRLYQYCGDVPPVELE AAYYAQRQRPAAG*
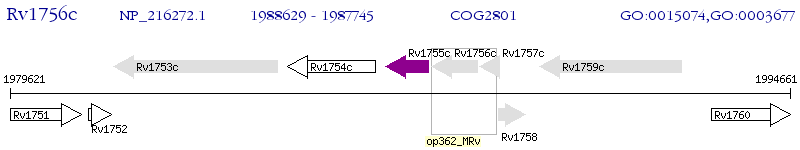
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1756c | - | - | 100% (294) | PUTATIVE TRANSPOSASE |
| M. tuberculosis H37Rv | Rv3475 | - | e-173 | 100.00% (294) | transposase IS6110 |
| M. tuberculosis H37Rv | Rv3380c | - | e-173 | 100.00% (294) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2838c | - | 1e-163 | 100.00% (278) | transposase |
| M. gilvum PYR-GCK | Mflv_0665 | - | 6e-85 | 51.69% (296) | integrase catalytic subunit |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2086c | - | 3e-29 | 55.96% (109) | putative transposase-like protein IS1601-D |
| M. marinum M | MMAR_2548 | - | 4e-81 | 51.06% (284) | transposase, ISMyma03_aa2 |
| M. avium 104 | MAV_5093 | - | 4e-83 | 53.26% (291) | transposase |
| M. smegmatis MC2 155 | MSMEG_1721 | - | 2e-79 | 49.66% (296) | IS1137, transposase orfB |
| M. thermoresistible (build 8) | TH_3317 | - | 2e-76 | 51.76% (284) | PUTATIVE Integrase, catalytic region |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5973 | - | 1e-57 | 54.03% (211) | integrase catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1756c|M.tuberculosis_H37Rv ----------------MRWGVESICTQLT-ELGVPIAPSTYYDHINRE-P
Mb2838c|M.bovis_AF2122/97 ---------------------------------MPIAPSTYYDHINRE-P
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5093|M.avium_104 --------MAFIDANRDEFGVEPICTVLRSAGLQVAPST-YYANKTRT-P
TH_3317|M.thermoresistible__bu -------MIAFIDAHRDQFGVELICRVLRAAIAGFLTSRGYRAAKARP-P
Mvan_5973|M.vanbaalenii_PYR-1 -----------------------------------MAPSTYYDVKSRC-P
Mflv_0665|M.gilvum_PYR-GCK -------MAGPDGGAGLRWGVEPMC-AVLSEHGIAISPSTYYEWIAKT-P
MMAR_2548|M.marinum_M ---------------------------MLTEHGIKISPSTYYEWMKRT-P
MSMEG_1721|M.smegmatis_MC2_155 MEFISAHQHMRVGVDGLKWGVESMC-AVLSEYGVTIAPSTYYAHRARQGP
Rv1756c|M.tuberculosis_H37Rv SRRELRDGELKEHISRVHAAN-----YGVYGARKVWLTLNREGIEVARCT
Mb2838c|M.bovis_AF2122/97 SRRELRDGELKEHISRVHAAN-----YGVYGARKVWLTLNREGIEVARCT
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5093|M.avium_104 SARAQRDAVMRPVITQLWEDN-----YRVYGARKLWKAARRAGHDIGRDQ
TH_3317|M.thermoresistible__bu SERAMRDELLIAELRTVHEQN-----FSVYGVKKMHHAMKRRGWRVGREQ
Mvan_5973|M.vanbaalenii_PYR-1 SARARRDAELGPALRVIWEDN-----YRVYGAVKLWKAARRGGHDVGRDQ
Mflv_0665|M.gilvum_PYR-GCK TRREIRDAELVEIITAAREGKRKGRFVQTLGSRKLWIWLRGQGHDVARCT
MMAR_2548|M.marinum_M SKQQLRDEAVTASIKEFRE---RNPLNKTLGSRKVWIKLRGDGMDVARCT
MSMEG_1721|M.smegmatis_MC2_155 SKADWADAQVIDAIWQLRQ---SQSLYRVLGARKTWIVLRTNGIDVSRCV
Rv1756c|M.tuberculosis_H37Rv VERLMTKLGLSGTTRGKARRTTIADPATARPADLVQRRFGPPAPNRLWVA
Mb2838c|M.bovis_AF2122/97 VERLMTKLGLSGTTRGKARRTTIADPATARPADLVQRRFGPPAPNRLWVA
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5093|M.avium_104 IARLMREAGIAGVRRGKRVRTTKPGPGAPRHPDLVGRDFTASAPNQLWVT
TH_3317|M.thermoresistible__bu TRRLMRKAGLRGVQRGKPVFTTISDPAHCRPADLVNRQFSAEAPDRLWVA
Mvan_5973|M.vanbaalenii_PYR-1 VARLMRSLGIEGVRRGKRVTTTRADPAAPRHPDLVKRQFTATAPNQLWVT
Mflv_0665|M.gilvum_PYR-GCK VERIMRENGWEGARYGSKHKTTVADSSHVRYPDLVDRRFYASAPNRLWVA
MMAR_2548|M.marinum_M VERLMQANRWQGARYGGAPKTTIQDEKAQRATDLVDRVFAAPAPNRLWVA
MSMEG_1721|M.smegmatis_MC2_155 VERVMREMGWRGACKRRRVRTTVADPAATRAPDRVRRNFVAGAPDRLWVA
Rv1756c|M.tuberculosis_H37Rv DLTYVSTWAGFAYVAFVTDAYARRILGWRVASTMATS-MVLDAIEQAIWT
Mb2838c|M.bovis_AF2122/97 DLTYVSTWAGFAYVAFVTDAYARRILGWRVASTMATS-MVLDAIEQAIWT
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5093|M.avium_104 DLTFVPTWAGVAYVCFLTDAFSRMIIGWRVASHMRTT-VVLDTIEMARWS
TH_3317|M.thermoresistible__bu DITFVRTWQGFCYTAFVTDACTKAIKGWAVAASMRTEDLPLEAFNHAVWH
Mvan_5973|M.vanbaalenii_PYR-1 DLTFVPTWVGVAYVCFIVDAFSRLIVGWRAASNMRTT-MVLDALEMARWS
Mflv_0665|M.gilvum_PYR-GCK DFTYVATWMGFVYVAFVIDAYSRRIVGWRAARSMTTD-LVLDAIEHAFFT
MMAR_2548|M.marinum_M DFTYCPTWAGMVYVAFVIDAFARRVIGWRAATSMKTA-LVIDALEHAFFT
MSMEG_1721|M.smegmatis_MC2_155 DFTYCRTRAGWAYTAFVTDVYARKIVGWKVATEMTQK-LVTDAIDHAIDT
Rv1756c|M.tuberculosis_H37Rv RQQEGVLDLKDVIHHTDRGSQYTSIRFSERLAEAGIQPSVGAVGSSYDNA
Mb2838c|M.bovis_AF2122/97 RQQEGVLDLKDVIHHTDRGSQYTSIRFSERLAEAGIQPSVGAVGSSYDNA
MAB_2086c|M.abscessus_ATCC_199 --------MPGLTCHSDAGSQFTSIRYGERLAEIGAVPSIGTIGDSYDNA
MAV_5093|M.avium_104 RGK----MLAGLRCHSDAGSQFTSIRYSERLAEIGAVPSIGTVGDSFDNA
TH_3317|M.thermoresistible__bu SDS----DLSELIHHSDRGSQYLSLTYTDRLAELGIAPSVGSRGDSYDNA
Mvan_5973|M.vanbaalenii_PYR-1 RGN----TLSGLRCHSDAGSQFTSIRYGERLAELGAVPSIGSVGDSFDNA
Mflv_0665|M.gilvum_PYR-GCK RTQDG-TSLRGLIAHSDAGSQYTSVAFTQRLIDEGVDPSVGSVGDALDNA
MMAR_2548|M.marinum_M RAQEGFTDLSGLVAHSDAGSQYTSIAFTTRLIDAGIDASVGSVADAYDNA
MSMEG_1721|M.smegmatis_MC2_155 RKRSGAASLDSLIHHSDAGSQYTAVAFTERLAAEGILPSVGSVGDSFDNA
: : *:* ***: :: : ** * .*:*: ..: ***
Rv1756c|M.tuberculosis_H37Rv LAETINGLYKTELIK---PGKPWRSIEDVELATARWVDWFNHRRLYQYCG
Mb2838c|M.bovis_AF2122/97 LAETINGLYKTELIK---PGKPWRSIEDVELATARWVDWFNHRRLYQYCG
MAB_2086c|M.abscessus_ATCC_199 L-ETVNGYYKAELIYGPVRTRPWKTVEDVELATLSWVYWHNTSRLHSYLN
MAV_5093|M.avium_104 LAETVNGYYKAELIYGPARPGPWKTVEDVELATLSWVHWHNTSRLHGYLN
TH_3317|M.thermoresistible__bu LAEAVNAAYKTELIN---RGKPWRSVDDVELATAGWVAWYNQERLHEALG
Mvan_5973|M.vanbaalenii_PYR-1 LAERAC------LID----------------CVSGWA-------------
Mflv_0665|M.gilvum_PYR-GCK LAETTVGSFKNEVIR---RQGPWRDVNQVELATAEWVVWFNTERPHEYLD
MMAR_2548|M.marinum_M LAETTVGSFKNELIR---RQGPWRDVDHVEIGTLNWVDWFNNERPHEYLA
MSMEG_1721|M.smegmatis_MC2_155 LAESVNSSYKTELID---RQPLYPGATELALGTAEWVAFYNRQRPNGYCQ
* * :* . *.
Rv1756c|M.tuberculosis_H37Rv DVPPVELEAAYYAQRQRPAAG-------
Mb2838c|M.bovis_AF2122/97 DVPPVELEAAYYAQRQRPAAG-------
MAB_2086c|M.abscessus_ATCC_199 DIPPTEFEAAFY-DAHGTDQPLAGIQ--
MAV_5093|M.avium_104 DVPPAEFEAAFY-ATKRTDHPLVEIQ--
TH_3317|M.thermoresistible__bu YVPPAEYEAALTGTSHPASQPTPALATN
Mvan_5973|M.vanbaalenii_PYR-1 ----------------------------
Mflv_0665|M.gilvum_PYR-GCK DFTPEAVEKLHYDQKLTPPKAG------
MMAR_2548|M.marinum_M DLTPTQAEALHYRHKSSLAAAG------
MSMEG_1721|M.smegmatis_MC2_155 DLTPDRAEALYYHRQRHPHAEEALR---