For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VADRLIVKGAREHNLRSVDLDLPRDALIVFTGLSGSGKSSLAFDTIFAEGQRRYVESLSAYARQFLGQMD KPDVDFIEGLSPAVSIDQKSTNRNPRSTVGTITEVYDYLRLLYARAGTPHCPTCGERVARQTPQQIVDQV LAMPEGTRFLVLAPVVRTRKGEFADLFDKLNAQGYSRVRVDGVVHPLTDPPKLKKQEKHDIEVVVDRLTV KAAAKRRLTDSVETALNLADGIVVLEFVDHELGAPHREQRFSEKLACPNGHALAVDDLEPRSFSFNSPYG ACPECSGLGIRKEVDPELVVPDPDRTLAQGAVAPWSNGHTAEYFTRMMAGLGEALGFDVDTPWRKLPAKA RKAILEGADEQVHVRYRNRYGRTRSYYADFEGVLAFLQRKMSQTESEQMKERYEGFMRDVPCPVCAGTRL KPEILAVTLAGESKGEHGAKSIAEVCELSIADCADFLNALTLGPREQAIAGQVLKEIRSRLGFLLDVGLE YLSLSRAAATLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIETLTRLRDLGNTLIVVEH DEDTIEHADWIVDIGPGAGEHGGRIVHSGPYDELLRNKDSITGAYLSGRESIEIPAIRRSVDPRRQLTVV GAREHNLRGIDVSFPLGVLTSVTGVSGSGKSTLVNDILAAVLANRLNGARQVPGRHTRVTGLDYLDKLVR VDQSPIGRTPRSNPATYTGVFDKIRTLFAATTEAKVRGYQPGRFSFNVKGGRCEACTGDGTIKIEMNFLP DVYVPCEVCQGARYNRETLEVHYKGKTVSEVLDMSIEEAAEFFEPIAGVHRYLRTLVDVGLGYVRLGQPA PTLSGGEAQRVKLASELQKRSTGRTVYILDEPTTGLHFDDIRKLLNVINGLVDKGNTVIVIEHNLDVIKT SDWIIDLGPEGGAGGGTVVAQGTPEDVAAVPASYTGKFLAEVVGGGASAATSRSNRRRNVSA
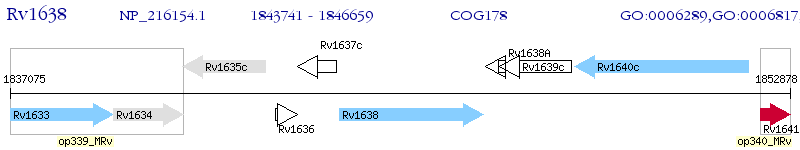
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1638 | uvrA | - | 100% (972) | excinuclease ABC subunit A |
| M. tuberculosis H37Rv | Rv1747 | - | 5e-09 | 31.40% (121) | transmembrane ATP-binding protein ABC transporter |
| M. tuberculosis H37Rv | Rv1859 | modC | 2e-05 | 33.93% (112) | molybdenum ABC transporter ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1664 | uvrA | 0.0 | 100.00% (972) | excinuclease ABC subunit A |
| M. gilvum PYR-GCK | Mflv_3548 | uvrA | 0.0 | 87.55% (972) | excinuclease ABC subunit A |
| M. leprae Br4923 | MLBr_01392 | uvrA | 0.0 | 90.54% (973) | excinuclease ABC subunit A |
| M. abscessus ATCC 19977 | MAB_2315 | uvrA | 0.0 | 86.57% (953) | excinuclease ABC subunit A |
| M. marinum M | MMAR_2443 | uvrA | 0.0 | 93.12% (974) | excinuclease ABC (subunit A-DNA-binding ATPase) UvrA |
| M. avium 104 | MAV_3135 | uvrA | 0.0 | 93.12% (974) | excinuclease ABC subunit A |
| M. smegmatis MC2 155 | MSMEG_3808 | uvrA | 0.0 | 88.17% (972) | excinuclease ABC subunit A |
| M. thermoresistible (build 8) | TH_1581 | uvrA | 0.0 | 88.07% (972) | excinuclease ABC, A subunit |
| M. ulcerans Agy99 | MUL_1622 | uvrA | 0.0 | 92.81% (974) | excinuclease ABC subunit A |
| M. vanbaalenii PYR-1 | Mvan_3339 | uvrA | 0.0 | 88.27% (972) | excinuclease ABC subunit A |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3548|M.gilvum_PYR-GCK MADRLVVKGAREHNLRGVDLDLPRDAMIVFTGLSGSGKSSLAFDTIFAEG
Mvan_3339|M.vanbaalenii_PYR-1 MADRLVVKGAREHNLRGVDLDLPRDAMIVFTGLSGSGKSSLAFDTIFAEG
MSMEG_3808|M.smegmatis_MC2_155 MADRLVVKGAREHNLRGVDLDLPRDALIVFTGLSGSGKSSLAFDTIFAEG
TH_1581|M.thermoresistible__bu LADRLIVKGAREHNLRSVDLDLPRDALIVFTGLSGSGKSSLAFDTIFAEG
Rv1638|M.tuberculosis_H37Rv MADRLIVKGAREHNLRSVDLDLPRDALIVFTGLSGSGKSSLAFDTIFAEG
Mb1664|M.bovis_AF2122/97 MADRLIVKGAREHNLRSVDLDLPRDALIVFTGLSGSGKSSLAFDTIFAEG
MMAR_2443|M.marinum_M MADRLIVKGAREHNLRGVDLDLPRDALIVFTGLSGSGKSSLAFDTIFAEG
MUL_1622|M.ulcerans_Agy99 MADRLIVKGAREHNLRGVDLDLPRDALIVFTGLSGSGKSSLAFDTIFAEG
MLBr_01392|M.leprae_Br4923 MADRLIVKGAREHNLRSVDLDLPRDSLIVFTGLSGSGKSSLAFDTIFAEG
MAV_3135|M.avium_104 MADRLIVKGAREHNLRGVDLDLPRDALIVFTGLSGSGKSSLAFDTIFAEG
MAB_2315|M.abscessus_ATCC_1997 MADRLIVRGAREHNLRGIDLDLPRDSLIVFTGLSGSGKSSLAFDTIFAEG
:****:*:********.:*******::***********************
Mflv_3548|M.gilvum_PYR-GCK QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
Mvan_3339|M.vanbaalenii_PYR-1 QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
MSMEG_3808|M.smegmatis_MC2_155 QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
TH_1581|M.thermoresistible__bu QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
Rv1638|M.tuberculosis_H37Rv QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
Mb1664|M.bovis_AF2122/97 QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
MMAR_2443|M.marinum_M QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
MUL_1622|M.ulcerans_Agy99 QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
MLBr_01392|M.leprae_Br4923 QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
MAV_3135|M.avium_104 QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
MAB_2315|M.abscessus_ATCC_1997 QRRYVESLSAYARQFLGQMDKPDVDFIEGLSPAVSIDQKSTNRNPRSTVG
**************************************************
Mflv_3548|M.gilvum_PYR-GCK TITEVYDYLRLLYARAGTPHCPVCGERIAKQTPQQIVDQVLAMDEGLRFQ
Mvan_3339|M.vanbaalenii_PYR-1 TITEVYDYLRLLYARAGTPHCPVCGERIARQTPQQIVDQVLAMDEGLRFQ
MSMEG_3808|M.smegmatis_MC2_155 TITEVYDYLRLLYARAGTPHCPVCGERIARQTPQQIVDQVLAMDEGLRFQ
TH_1581|M.thermoresistible__bu TITEVYDYLRLLYARAGTPHCPVCGERIARQTPQQIVDQVLAMDEGLRFQ
Rv1638|M.tuberculosis_H37Rv TITEVYDYLRLLYARAGTPHCPTCGERVARQTPQQIVDQVLAMPEGTRFL
Mb1664|M.bovis_AF2122/97 TITEVYDYLRLLYARAGTPHCPTCGERVARQTPQQIVDQVLAMPEGTRFL
MMAR_2443|M.marinum_M TITEVYDYLRLLYARAGTPHCPTCGERIARQTPQQIVDQVLAMAEGTRFL
MUL_1622|M.ulcerans_Agy99 TITEVYDYLRLLYARAGTPHCPTCGERIARQTPQQIVDQVLAMAEGTRFL
MLBr_01392|M.leprae_Br4923 TITEVYDYLRLLYARAGTPHCPICGEQISRQTPQQIVDQVLVLPEGTRFL
MAV_3135|M.avium_104 TITEVYDYLRLLYARAGTPHCPVCGERIARQTPQQIVDQVLAMPEGTRFL
MAB_2315|M.abscessus_ATCC_1997 TITEVYDYLRLLYARAGTPHCPVCGEKIAKQTPQQIVDQVLDMEEGLRFQ
********************** ***::::*********** : ** **
Mflv_3548|M.gilvum_PYR-GCK VLAPVVRTRKGEFVDLFDKLNTQGYSRVRVDGVVYPLTDPPKLKKQEKHD
Mvan_3339|M.vanbaalenii_PYR-1 VLAPVVRTRKGEFVDLFDKLNSQGYSRVRVDGVVHPLTDPPKLKKQEKHD
MSMEG_3808|M.smegmatis_MC2_155 VLAPVVRTRKGEFVDLFEKLNSQGYSRVRVDGVVYPLTDPPKLKKQEKHD
TH_1581|M.thermoresistible__bu VLAPVVRTRKGEFVDLFDKLNSQGYSRVRVDGVVYPLTDPPKLKKQEKHD
Rv1638|M.tuberculosis_H37Rv VLAPVVRTRKGEFADLFDKLNAQGYSRVRVDGVVHPLTDPPKLKKQEKHD
Mb1664|M.bovis_AF2122/97 VLAPVVRTRKGEFADLFDKLNAQGYSRVRVDGVVHPLTDPPKLKKQEKHD
MMAR_2443|M.marinum_M VLAPVVRTRKGEFADLFDKLNAQGYSRVRVDGVVHPLTDPPKLKKQEKHD
MUL_1622|M.ulcerans_Agy99 VLAPVVRTRKGEFADLFDKLNAQGYSRVRVDGVVHPLTDPPKLKKQEKHD
MLBr_01392|M.leprae_Br4923 VLAPVVRTRKGEFADLFDKLNAQGYSRVRVDDVVYPLSDPPKLKKQEKHD
MAV_3135|M.avium_104 VLAPVVRTRKGEFADLFEKLNAQGYSRVRVDGVVHPLTDPPKLKKQEKHD
MAB_2315|M.abscessus_ATCC_1997 VLAPVVRTRKGEFVDLFEQLNSQGYSRIRVDGVVHSLTDPPKLKKQEKHD
*************.***::**:*****:***.**:.*:************
Mflv_3548|M.gilvum_PYR-GCK IEVVVDRLTVKASSKQRLTDSIETALNLADGIVVLEFVD-REDDHPHREQ
Mvan_3339|M.vanbaalenii_PYR-1 IEVVVDRLTVKATAKQRLTDSVETALNLADGIVVLEFVD-REDDHPHREQ
MSMEG_3808|M.smegmatis_MC2_155 IEVVVDRLTVKASAKQRLTDSIETALNLADGIVVLEFVD-REDDHPHREQ
TH_1581|M.thermoresistible__bu IEVVVDRLTVKPSAKQRLTDSIETALNLADGIVVLDFVD-RDEDDPHREQ
Rv1638|M.tuberculosis_H37Rv IEVVVDRLTVKAAAKRRLTDSVETALNLADGIVVLEFVD-HELGAPHREQ
Mb1664|M.bovis_AF2122/97 IEVVVDRLTVKAAAKRRLTDSVETALNLADGIVVLEFVD-HELGAPHREQ
MMAR_2443|M.marinum_M IEVVVDRLTVKAAAKQRLTDSVETALNLADGIVVLEFPDDHDEHDHPREQ
MUL_1622|M.ulcerans_Agy99 IEVVVDRLTVKAAAKQRLTDSVETALNLADGIVVLEFPDDHDEHDHPREQ
MLBr_01392|M.leprae_Br4923 IEVVVDRLTVKIAAKQRLTDSVETALNLADGIVVLEFPDDHDERGHPREQ
MAV_3135|M.avium_104 IEVVVDRLTVKASAKQRLTDSVETALNLADGIVVLEFVD-HEHDAHNREQ
MAB_2315|M.abscessus_ATCC_1997 IEVVIDRLSVKASSKQRLTDSVETALHLADGIVVLEFVD-ADEDSPQRER
****:***:** ::*:*****:****:********:* * : **:
Mflv_3548|M.gilvum_PYR-GCK RFSEKLACPNGHPLAVDDLEPRSFSFNSPYGACPECLGLGIRKEVDPDLV
Mvan_3339|M.vanbaalenii_PYR-1 RFSEKLACPNGHPLAVDDLEPRSFSFNSPYGACPECTGLGIRKEVDPDLV
MSMEG_3808|M.smegmatis_MC2_155 RFSEKLACPNGHPLAVDDLEPRSFSFNSPYGACPECTGLGIRKEVDPDLV
TH_1581|M.thermoresistible__bu RFSEKLACPNGHPLAVDDLEPRSFSFNSPYGACPECAGLGIRKEVDVDLV
Rv1638|M.tuberculosis_H37Rv RFSEKLACPNGHALAVDDLEPRSFSFNSPYGACPECSGLGIRKEVDPELV
Mb1664|M.bovis_AF2122/97 RFSEKLACPNGHALAVDDLEPRSFSFNSPYGACPECSGLGIRKEVDPELV
MMAR_2443|M.marinum_M RFSEKLACPNGHPLAVDDLEPRSFSFNSPYGACPECSGLGIRKEVDPDLV
MUL_1622|M.ulcerans_Agy99 RFSEKLACPNGHPLAVDDLEPRSFSFNSPYGACPECSGLGIRKEVDPDLV
MLBr_01392|M.leprae_Br4923 CFSEKLACPNGHPLAVDDLEPRSFSFNSPYGACPECVGLGIRKEIDSDLV
MAV_3135|M.avium_104 RFSEKLACPNGHALAVDDLEPRSFSFNSPYGACPECSGLGIRKEVDPDLV
MAB_2315|M.abscessus_ATCC_1997 RFSEKLACPNGHPLAVDDLEPRSFSFNSPYGACPECAGLGIKKEVDPDLV
***********.*********************** ****:**:* :**
Mflv_3548|M.gilvum_PYR-GCK VPDPDLTLAEGAIAPWSMGNTADYFTRMLSGLGDQLGFDVNTPWKKLPAK
Mvan_3339|M.vanbaalenii_PYR-1 VPDPDLTLAEGAIAPWSMGHTAEYFTRMLSGLGDQLGFDVNTPWKKLPAK
MSMEG_3808|M.smegmatis_MC2_155 VPDPELTLAEGAVAPWSVGQSAEYFTRMLAGLGEEMGFDVNTPWKKLPAK
TH_1581|M.thermoresistible__bu VPDPDLTLAEGAIAPWSMGQTAEYFTRMLAGLGEAMGFDVDTPWRKLPAK
Rv1638|M.tuberculosis_H37Rv VPDPDRTLAQGAVAPWSNGHTAEYFTRMMAGLGEALGFDVDTPWRKLPAK
Mb1664|M.bovis_AF2122/97 VPDPDRTLAQGAVAPWSNGHTAEYFTRMMAGLGEALGFDVDTPWRKLPAK
MMAR_2443|M.marinum_M VPDPDRTLAEGAVAPWSTGHTAEYFTRMLAGLGDALGFDVDTPWRKLPAK
MUL_1622|M.ulcerans_Agy99 VPDPDRTLAEGAVAPWSTGHTAEYFTRMLAGLGDALGFDVDTPWRKLPAK
MLBr_01392|M.leprae_Br4923 VPDPDRTLAEGAVAPWSAGRTAEYFTRMMAGLGEKLGFDVDTPWRKLPAK
MAV_3135|M.avium_104 VPDPERTLAEGAVAPWSTGHTAEYFTRMMAGLGDELGFDVDTPWRKLPAK
MAB_2315|M.abscessus_ATCC_1997 VPDPDMTLAEGAIAPWSMGQNADYFVRLMAGLGDALGFDVDTPWKKLPAA
****: ***:**:**** *..*:**.*:::***: :****:***:****
Mflv_3548|M.gilvum_PYR-GCK ARKAILEGCDEQVHVRYKNRYGRTRSYYADFEGVMAFLQRRMEQTDSEQT
Mvan_3339|M.vanbaalenii_PYR-1 ARRAILEGCDEQVHVRYKNRYGRTRSYYADFEGVMAFLQRRMEQTDSEQM
MSMEG_3808|M.smegmatis_MC2_155 ARRAILEGCDHQVHVRYKNRYGRTRSYYADFEGVMAFLQRRMEQTDSEQM
TH_1581|M.thermoresistible__bu VRKAILEGCDEQVHVRYKNRYGRVRSYYADFEGVMAYLHRRMEQTDSEQM
Rv1638|M.tuberculosis_H37Rv ARKAILEGADEQVHVRYRNRYGRTRSYYADFEGVLAFLQRKMSQTESEQM
Mb1664|M.bovis_AF2122/97 ARKAILEGADEQVHVRYRNRYGRTRSYYADFEGVLAFLQRKMSQTESEQM
MMAR_2443|M.marinum_M ARKAILEGADEQVHVRYRNRYGRTRSYYADFEGVLAFLQRKMAQTESEQM
MUL_1622|M.ulcerans_Agy99 ARKAILEGADEQVHVRYRNRYGRTRSYYADFEGVLAFLQRKMAQTESEQM
MLBr_01392|M.leprae_Br4923 ARKAILEGSDHQVHVQYHNRYGRPRSYYVDFEGVLTFLQRKMEQTESEQM
MAV_3135|M.avium_104 ARKAILEGSDHQVHVRYRNRYGRTRSYYADFEGVLAFLQRKMAQTESEQM
MAB_2315|M.abscessus_ATCC_1997 AKKAILDGSSEQVHVRYKNRYGRTRSYYAEFEGVLAFLQRRMEQTESEWA
.::***:*...****:*:***** ****.:****:::*:*:* **:**
Mflv_3548|M.gilvum_PYR-GCK KERLEGFMRDVPCPECDGTRLKPEILAVTMTA---GTFGAKSIAEVSELS
Mvan_3339|M.vanbaalenii_PYR-1 KERLEGFMRDVPCPECDGTRLKPEILAVTMTA---GNFGAKSIAEVAELS
MSMEG_3808|M.smegmatis_MC2_155 KERYEGFMRDIPCPECNGTRLKPEILAVTLSA---GDFGAKSIAQVAELS
TH_1581|M.thermoresistible__bu KERLEGFMRDVPCPECGGTRLKPEILAVTLSA---GEFGEKSIAEVADLS
Rv1638|M.tuberculosis_H37Rv KERYEGFMRDVPCPVCAGTRLKPEILAVTLAGESKGEHGAKSIAEVCELS
Mb1664|M.bovis_AF2122/97 KERYEGFMRDVPCPVCAGTRLKPEILAVTLAGESKGEHGAKSIAEVCELS
MMAR_2443|M.marinum_M KERYEGFMRDVPCPVCDGARLKPEILSVTLAA---GEQDAKSIAEVCELS
MUL_1622|M.ulcerans_Agy99 KERYEGFMRDVPCPVCDGARLKPEILSVTLAA---GEQDAKSIAEVCELS
MLBr_01392|M.leprae_Br4923 KERYEGFMRNIPCPVCQGTRLKPEILAVTLAA---GERGAKSIAEVCELS
MAV_3135|M.avium_104 KERYEGFMRDVPCPVCEGTRLKPEILAVTLAA---GRHGKKSIAEVCELS
MAB_2315|M.abscessus_ATCC_1997 KERYEGFMRDIPCPVCEGTRLKPEILSVTLTA---GEHGTKSIAEVCDLS
*** *****::*** * *:*******:**::. * . ****:*.:**
Mflv_3548|M.gilvum_PYR-GCK IADCAAFLNALTLGTREAAIAGQVLKEIQSRLGFLLDVGLDYLSLSRAAG
Mvan_3339|M.vanbaalenii_PYR-1 IADCAEFLNALTLGTREAAIAGQVLKEIQSRLGFLLDVGLDYLSLSRAAG
MSMEG_3808|M.smegmatis_MC2_155 IADCADFLNSLTLGPREQAIAGQVLKEIQSRLGFLLDVGLDYLSLSRAAA
TH_1581|M.thermoresistible__bu IADCAEFLNALTLGPREQAIAGQVLKEIQSRLGFLLDVGLDYLSLSRAAA
Rv1638|M.tuberculosis_H37Rv IADCADFLNALTLGPREQAIAGQVLKEIRSRLGFLLDVGLEYLSLSRAAA
Mb1664|M.bovis_AF2122/97 IADCADFLNALTLGPREQAIAGQVLKEIRSRLGFLLDVGLEYLSLSRAAA
MMAR_2443|M.marinum_M ISDCADFLNALTLGAREQAIAGQVLKEIQSRLGFLLDVGLEYLSLSRAAA
MUL_1622|M.ulcerans_Agy99 ISDCADFLNALTLGAREQAIAGQVLKEIQSRLGFLLDVGLEYLSLSRAAA
MLBr_01392|M.leprae_Br4923 IADCSAFLNALILGVREQAIAGQVLKEIQSRLGFLLDVGLEYLSLSRAAA
MAV_3135|M.avium_104 ISDCAEFLNTLTLGPREQAIAGQVLKEIQSRLGFLLDVGLEYLSLSRAAA
MAB_2315|M.abscessus_ATCC_1997 IADCAQFLGALTLGHREQAIAGQVLKEVQFRLGFLLDVGLQYLSLSRAAG
*:**: **.:* ** ** *********:: **********:********.
Mflv_3548|M.gilvum_PYR-GCK TLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIDTLVRLR
Mvan_3339|M.vanbaalenii_PYR-1 TLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIDTLVRLR
MSMEG_3808|M.smegmatis_MC2_155 TLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIDTLVRLR
TH_1581|M.thermoresistible__bu TLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIDTLVRLR
Rv1638|M.tuberculosis_H37Rv TLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIETLTRLR
Mb1664|M.bovis_AF2122/97 TLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIETLTRLR
MMAR_2443|M.marinum_M TLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIETLTRLR
MUL_1622|M.ulcerans_Agy99 TLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIETLTRLR
MLBr_01392|M.leprae_Br4923 TLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIETLTRLR
MAV_3135|M.avium_104 TLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIETLTRLR
MAB_2315|M.abscessus_ATCC_1997 TLSGGEAQRIRLATQIGSGLVGVLYVLDEPSIGLHQRDNRRLIETLTRLR
*******************************************:**.***
Mflv_3548|M.gilvum_PYR-GCK DLGNTLIVVEHDLDTIAHADWVVDIGPAAGEHGGQIVHSGTYADLLTNPE
Mvan_3339|M.vanbaalenii_PYR-1 DLGNTLIVVEHDLDTIAHADWVVDIGPAAGEHGGQIVHSGTYEDLLRNPN
MSMEG_3808|M.smegmatis_MC2_155 DLGNTLIVVEHDLDTIAHADWVVDIGPAAGEHGGQIVHSGTYDDLLRNPE
TH_1581|M.thermoresistible__bu DLGNTLIVVEHDLDTIAHADWVVDIGPAAGEHGGRIVHSGPYADLLANPD
Rv1638|M.tuberculosis_H37Rv DLGNTLIVVEHDEDTIEHADWIVDIGPGAGEHGGRIVHSGPYDELLRNKD
Mb1664|M.bovis_AF2122/97 DLGNTLIVVEHDEDTIEHADWIVDIGPGAGEHGGRIVHSGPYDELLRNKD
MMAR_2443|M.marinum_M DLGNTLIVVEHDEDTIEHADWVVDIGPGAGEHGGSIVHSGPYQELLRNQS
MUL_1622|M.ulcerans_Agy99 DLGNTLIVVEHDEDTIEHADWVVDIGPGAGEHGGSIVHSGPYQELLRNQS
MLBr_01392|M.leprae_Br4923 ALGNTLIVVEHDEDTIAHADWVVDIGPGAGEHGGQIVHNGTYRELLANKD
MAV_3135|M.avium_104 DLGNTLIVVEHDEDTIAHADWIVDIGPRAGEHGGQIVHSGTYAELLANQD
MAB_2315|M.abscessus_ATCC_1997 DLGNTLIVVEHDEDTIKHADWVVDIGPAAGEHGGQVVHSGTYAELLKNKN
*********** *** ****:***** ****** :**.*.* :** * .
Mflv_3548|M.gilvum_PYR-GCK SITGAYLSGKEEIEVPTMRRVVDKKRQLTVVGAREHNLREIDVSFPLGVL
Mvan_3339|M.vanbaalenii_PYR-1 SITGAYLSGKEEIEVPDMRRVVDRKRQLTVVGAREHNLREIDVAFPLGVL
MSMEG_3808|M.smegmatis_MC2_155 SLTGAYLSGKESIEVPAIRRPVDKKRQITVVGARENNLKEIDVAFPLGVL
TH_1581|M.thermoresistible__bu SLTGEYLSGKKSIEVPAMRRPVDRKRQLTVVGARENNLRDIDVSFPLGVL
Rv1638|M.tuberculosis_H37Rv SITGAYLSGRESIEIPAIRRSVDPRRQLTVVGAREHNLRGIDVSFPLGVL
Mb1664|M.bovis_AF2122/97 SITGAYLSGRESIEIPAIRRSVDPRRQLTVVGAREHNLRGIDVSFPLGVL
MMAR_2443|M.marinum_M SITGAFLSGREGIEIPTSRRSVDSRRQLTVVGAREHNLRGIDVAFPLGVL
MUL_1622|M.ulcerans_Agy99 SITGAFLSGREGIEIPTSRRSIDSRRQLTVVGAREHNLRGLDVAFPLGVL
MLBr_01392|M.leprae_Br4923 SITGAYLSGRESIATPTRRRSVDRKRELTVVGAREHNLRGIDVSFPLGVL
MAV_3135|M.avium_104 SITGAYLSGKESIEMPAIRRPVDRRRQLTVVGAREHNLRGIDVSFPLGVL
MAB_2315|M.abscessus_ATCC_1997 SVTGSYLSGAESIPLPLIRRVIDPKRQVTVVGAREHNLKDIDVSFPLGVL
*:** :*** : * * ** :* :*::*******:**: :**:******
Mflv_3548|M.gilvum_PYR-GCK TSVTGVSGSGKSTLVNDILASVLANKLNGARLVPGRHTRINGLDHLDKLV
Mvan_3339|M.vanbaalenii_PYR-1 TSVTGVSGSGKSTLVNDILASVLANKLNGARLVPGRHTRVNGLDHLDKLV
MSMEG_3808|M.smegmatis_MC2_155 TSVTGVSGSGKSTMVNDILATVLANKLNGARLVPGRHTRVNGLDQLDKLV
TH_1581|M.thermoresistible__bu TAVTGVSGSGKSTLVNDILAAALANKLNGARLVPGRHTRVNGLGEVDKLV
Rv1638|M.tuberculosis_H37Rv TSVTGVSGSGKSTLVNDILAAVLANRLNGARQVPGRHTRVTGLDYLDKLV
Mb1664|M.bovis_AF2122/97 TSVTGVSGSGKSTLVNDILAAVLANRLNGARQVPGRHTRVTGLDYLDKLV
MMAR_2443|M.marinum_M TSVTGVSGSGKSTLVNDILAAVLANRLNGARQVPGRHTRITGLDHLDKLV
MUL_1622|M.ulcerans_Agy99 TSVTGVSGSGKSTLVNDILAAVLANRLNGARQVPGRHTRITGLDHLDKLV
MLBr_01392|M.leprae_Br4923 TSVTGVSGSGKSTLVNDILAAVLANRLNGARQVPGRHTRVTGLEHLDKLV
MAV_3135|M.avium_104 TSVTGVSGSGKSTLVNDILATVLANRLNGARLVPGRHTRVTGLDHLDKLV
MAB_2315|M.abscessus_ATCC_1997 TSVTGVSGSGKSTLVNDILATVLANKLNGARQVPGRHTRVTGLDKVDKLV
*:***********:******:.***:***** *******:.** :****
Mflv_3548|M.gilvum_PYR-GCK RVDQSPIGRTPRSNPATYTGVFDKIRTLFAATTEAKVRGYQPGRFSFNVK
Mvan_3339|M.vanbaalenii_PYR-1 RVDQSPIGRTPRSNPATYTGVFDKIRTLFAATTEAKVRGYQPGRFSFNVK
MSMEG_3808|M.smegmatis_MC2_155 RVDQSPIGRTPRSNPATYTGVFDKIRSLFAATTEAKVRGYQPGRFSFNVK
TH_1581|M.thermoresistible__bu RVDQSPIGRTPRSNPATYTGVFDKIRTLFAATTEAKVRGYQPGRFSFNVK
Rv1638|M.tuberculosis_H37Rv RVDQSPIGRTPRSNPATYTGVFDKIRTLFAATTEAKVRGYQPGRFSFNVK
Mb1664|M.bovis_AF2122/97 RVDQSPIGRTPRSNPATYTGVFDKIRTLFAATTEAKVRGYQPGRFSFNVK
MMAR_2443|M.marinum_M RVDQSPIGRTPRSNPATYTGVFDKIRTLFAATTEAKVRGYQPGRFSFNVK
MUL_1622|M.ulcerans_Agy99 RVDQSPIGRTPRSNPATYTGVFDKIRTLFAATTEAKVRGYQPGRFSFNVK
MLBr_01392|M.leprae_Br4923 RVDQSPIGRTPRSNPATYTGVFDKIRILFAATTEAKVRGYQPGRFSFNVK
MAV_3135|M.avium_104 RVDQSPIGRTPRSNPATYTGVFDKIRTLFAATTEAKVRGYQPGRFSFNVK
MAB_2315|M.abscessus_ATCC_1997 RVDQSPIGRTPRSNPATYTGVFDKIRTLFAATTEAKVRGYQPGRFSFNVK
************************** ***********************
Mflv_3548|M.gilvum_PYR-GCK GGRCEACSGDGTIKIEMNFLPDVYVPCEVCQGARYNRETLEVHYKGKTIA
Mvan_3339|M.vanbaalenii_PYR-1 GGRCEACSGDGTIKIEMNFLPDVYVPCEVCQGARYNRETLEVHYKGKTIA
MSMEG_3808|M.smegmatis_MC2_155 GGRCEACSGDGTIKIEMNFLPDVYVPCEVCHGARYNRETLEVHYKGKTIS
TH_1581|M.thermoresistible__bu GGRCEACSGDGTLKIEMNFLPDVYVPCEVCHGARYNRETLEVHYKGKTIA
Rv1638|M.tuberculosis_H37Rv GGRCEACTGDGTIKIEMNFLPDVYVPCEVCQGARYNRETLEVHYKGKTVS
Mb1664|M.bovis_AF2122/97 GGRCEACTGDGTIKIEMNFLPDVYVPCEVCQGARYNRETLEVHYKGKTVS
MMAR_2443|M.marinum_M GGRCEACTGDGTIKIEMNFLPDVYVPCEVCQGARYNRETLEVHYKGKTIS
MUL_1622|M.ulcerans_Agy99 GGRCEACTGDGTIKIEMNFLPDVYVPCEVCQGARYNRETLEVHYNGKTIS
MLBr_01392|M.leprae_Br4923 GGRCEACTGDGTIKIEMNFLPDVYVPCEVCQGARYNRETLEVHYKGKAIS
MAV_3135|M.avium_104 GGRCEACTGDGTIKIEMNFLPDVYVPCEVCHGARYNRETLEVHYKGKTIS
MAB_2315|M.abscessus_ATCC_1997 GGRCEACTGDGTIKIEMNFLPDVYVPCEVCHGARYNRETLEVHYKGKTIA
*******:****:*****************:*************:**:::
Mflv_3548|M.gilvum_PYR-GCK EVLDMSIEEAAEFFAPISSIHRYLKTLVDVGLGYVRLGQPAPTLSGGEAQ
Mvan_3339|M.vanbaalenii_PYR-1 EVLDMSIEEAAEFFAPISSIHRYLKTLVDVGLGYVRLGQPAPTLSGGEAQ
MSMEG_3808|M.smegmatis_MC2_155 EVLDMSIEEATEFFEPISSIHRYLKTLVDVGLGYVRLGQPAPTLSGGEAQ
TH_1581|M.thermoresistible__bu EVLDMSIEEAAEFFAPISSIHRYLQTLVDVGLGYVRLGQPAPTLSGGEAQ
Rv1638|M.tuberculosis_H37Rv EVLDMSIEEAAEFFEPIAGVHRYLRTLVDVGLGYVRLGQPAPTLSGGEAQ
Mb1664|M.bovis_AF2122/97 EVLDMSIEEAAEFFEPIAGVHRYLRTLVDVGLGYVRLGQPAPTLSGGEAQ
MMAR_2443|M.marinum_M EVLDMSIEEAADFFEPISGIHRYLRTLVDVGLGYVRLGQPAPTLSGGEAQ
MUL_1622|M.ulcerans_Agy99 EVLDMSIEEAADFFEPISGIHRYLRTLVDVGLGYVRLGQPAPTLSGGEAQ
MLBr_01392|M.leprae_Br4923 EVLDMSIEEAAEFFEPIIGINRYLRTLVDVGLGYVRLGQPAPTLSGGEAQ
MAV_3135|M.avium_104 EVLDMSIEEAAEFFEPITGIHRYLRTLVDVGLGYVRLGQPAPTLSGGEAQ
MAB_2315|M.abscessus_ATCC_1997 QVLDMPIEEAAEFFEPITSIHRYLKTLVEVGLGYVRLGQPAPTLSGGEAQ
:****.****::** ** .::***:***:*********************
Mflv_3548|M.gilvum_PYR-GCK RVKLASELQKRSTGRTIYILDEPTTGLHFEDIRKLLTVINGLVDKGNTVI
Mvan_3339|M.vanbaalenii_PYR-1 RVKLASELQKRSTGRTIYILDEPTTGLHFEDIRKLLTVINGLVDKGNTVI
MSMEG_3808|M.smegmatis_MC2_155 RVKLAAELQKRSTGRTIYILDEPTTGLHFEDIRKLLKVINGLVDKGNTVI
TH_1581|M.thermoresistible__bu RVKLASELQKRSTGRTVYILDEPTTGLHFEDIRKLLGVINGLVDKGNTVI
Rv1638|M.tuberculosis_H37Rv RVKLASELQKRSTGRTVYILDEPTTGLHFDDIRKLLNVINGLVDKGNTVI
Mb1664|M.bovis_AF2122/97 RVKLASELQKRSTGRTVYILDEPTTGLHFDDIRKLLNVINGLVDKGNTVI
MMAR_2443|M.marinum_M RVKLASELQKRSTGRTIYILDEPTTGLHFDDIRKLLNVISGLVDKGNTVV
MUL_1622|M.ulcerans_Agy99 RVKLASELQKRSTGRTIYILDEPTTGLHFDDIRKLLNVISGLVDKGNTVV
MLBr_01392|M.leprae_Br4923 RVKLASELQKRSTGRTIYILDEPTTGLHFDDIRKLLNVINGLVDKDNTVI
MAV_3135|M.avium_104 RVKLAAELQKRSTGRTIYILDEPTTGLHFDDIRKLLKVINGLVDKGNTVI
MAB_2315|M.abscessus_ATCC_1997 RVKLAAELQKRSTGKTIYILDEPTTGLHFEDIRKLLTVINGLVDKGNTVI
*****:********:*:************:****** **.*****.***:
Mflv_3548|M.gilvum_PYR-GCK VIEHNLDVIKTSDWIIDMGPEGGSGGGTVVATGAPEDVAMNPDSYTGHFL
Mvan_3339|M.vanbaalenii_PYR-1 VIEHNLDVIKTSDWVVDMGPEGGSGGGTVVATGTPEDVAANPDSYTGHFL
MSMEG_3808|M.smegmatis_MC2_155 VIEHNLDVIKTSDWIIDMGPEGGAGGGTVVAQGTPEDVAANPDSYTGKFL
TH_1581|M.thermoresistible__bu VIEHNLDVIKTADWIIDMGPEGGNGGGTVVAQGTPEDVAANPDSYTGQFL
Rv1638|M.tuberculosis_H37Rv VIEHNLDVIKTSDWIIDLGPEGGAGGGTVVAQGTPEDVAAVPASYTGKFL
Mb1664|M.bovis_AF2122/97 VIEHNLDVIKTSDWIIDLGPEGGAGGGTVVAQGTPEDVAAVPASYTGKFL
MMAR_2443|M.marinum_M VIEHNLDVIKTSDWIIDMGPEGGAGGGTVVAQGAPEDIAAVPESYTGKFL
MUL_1622|M.ulcerans_Agy99 VIEHNLDVIKTSDWIIDMGPEGGAGGGTVVAQGAPEDIAAVPESYTGKFL
MLBr_01392|M.leprae_Br4923 VIEHNLDVIKTSDWIVDMGPEGGAQGGTVVAEGTPEDVAAVPESYTGKFL
MAV_3135|M.avium_104 VIEHNLDVIKTSDWIVDMGPEGGAEGGTVVAQGTPEDVAAVPESYTGKFL
MAB_2315|M.abscessus_ATCC_1997 VIEHNLDVIKTSDWIIDMGPEGGSGGGTVVAEGTPESVAQTAGSYTGEFL
***********:**::*:***** ****** *:**.:* . ****.**
Mflv_3548|M.gilvum_PYR-GCK AETLGMN----------RPKPK------ARARRKVSA--
Mvan_3339|M.vanbaalenii_PYR-1 AETLGVA----------RPKPK------SRRRSKVSA--
MSMEG_3808|M.smegmatis_MC2_155 AELLDVP----------TPK---------RKRRKVSA--
TH_1581|M.thermoresistible__bu AEILDVP----------APT---------RKRRKVQVSA
Rv1638|M.tuberculosis_H37Rv AEVVGGGA------SAATSR--------SNRRRNVSA--
Mb1664|M.bovis_AF2122/97 AEVVGGGA------SAATSR--------SNRRRNVSA--
MMAR_2443|M.marinum_M AEVLAARR------PSTTARR-------TNRRRKVSA--
MUL_1622|M.ulcerans_Agy99 AEVLAARR------PSTTARR-------TNRRRKVSA--
MLBr_01392|M.leprae_Br4923 AEVVGAG--------CAPART-------PRRRRTVTA--
MAV_3135|M.avium_104 AEVIGRGG------AAAGAPRR------SSRRRKATA--
MAB_2315|M.abscessus_ATCC_1997 VDLLPRPVRNGKAPAKAPAKSTGKTLAKARTRKVVTPV-
.: : . * .