For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPIPAPSPDARAVVTGASQNIGAALATELAARGHHLIVTARREDVLTELAARLADKYRVTVDVRPADLAD PQERSKLADELAARPISILCANAGTATFGPIASLDLAGEKTQVQLNAVAVHDLTLAVLPGMIERKAGGIL ISGSAAGNSPIPYNATYAATKAFVNTFSESLRGELRGSGVHVTVLAPGPVRTELPDASEASLVEKLVPDF LWISTEHTARVSLNALERNKMRVVPGLTSKAMSVASQYAPRAIVAPIVGAFYKRLGGS
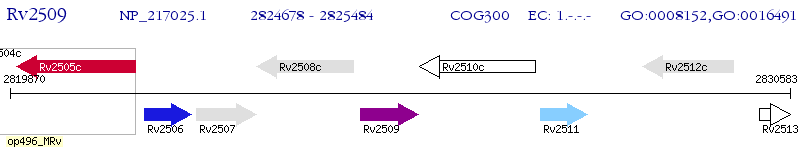
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2509 | - | - | 100% (268) | PROBABLE SHORT-CHAIN TYPE DEHYDROGENASE/REDUCTASE |
| M. tuberculosis H37Rv | Rv1544 | - | 3e-52 | 43.07% (267) | ketoacyl reductase |
| M. tuberculosis H37Rv | Rv0765c | - | 2e-23 | 33.94% (221) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2537 | - | 1e-147 | 100.00% (268) | putative short-chain type dehydrogenase/reductase |
| M. gilvum PYR-GCK | Mflv_2545 | - | 1e-113 | 76.40% (267) | short-chain dehydrogenase/reductase SDR |
| M. leprae Br4923 | MLBr_00429 | - | 1e-131 | 88.76% (267) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_1537c | - | 1e-114 | 78.20% (266) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_3856 | - | 1e-136 | 91.76% (267) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_1666 | - | 1e-131 | 87.27% (267) | short-chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4722 | - | 1e-117 | 79.03% (267) | short-chain dehydrogenase |
| M. thermoresistible (build 8) | TH_1383 | - | 1e-118 | 79.40% (267) | PROBABLE SHORT-CHAIN TYPE DEHYDROGENASE/REDUCTASE |
| M. ulcerans Agy99 | MUL_3788 | - | 1e-135 | 91.01% (267) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_4098 | - | 1e-114 | 76.78% (267) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2509|M.tuberculosis_H37Rv MPIPAPSPDARAVVTGASQNIGAALATELAARGHHLIVTARREDVLTELA
Mb2537|M.bovis_AF2122/97 MPIPAPSPDARAVVTGASQNIGAALATELAARGHHLIVTARREDVLTELA
MLBr_00429|M.leprae_Br4923 MPIPAPSPEARAVVTGASQNIGEALATELAAHGHSLIVIARREDVLVDLA
MMAR_3856|M.marinum_M MPIPAPSPDARAVVTGASQNIGEALATELAARGHNLIVTARRESLLNELA
MUL_3788|M.ulcerans_Agy99 MPIPAPSPDARAVVTGASQNIGEALATELAARGHNLIVTARRESLLNELA
MAV_1666|M.avium_104 MPIPAPSPEARAVVTGASQNIGEALATELAARGHHLIVTARREDLLKDLA
MAB_1537c|M.abscessus_ATCC_199 MPIPAPSPDARAVVTGASQGIGAALAEELAARGHNLIITARRGEILNDLA
Mflv_2545|M.gilvum_PYR-GCK MPVPAPSADARAVVTGASQNIGEALAVELARRGHHLIITARREEVLNALA
Mvan_4098|M.vanbaalenii_PYR-1 MPVPAPSEDARAVVTGASQNIGEALAVELAERGHHLIITARREEVLNALA
MSMEG_4722|M.smegmatis_MC2_155 MPVPAPSPDARAVVTGASQNIGEALATELAARGHHLIITARREEVLTSLA
TH_1383|M.thermoresistible__bu MPVPEPTPDARAVITGASQNIGAALATELAARGHNLIVTARRAEVLTELA
**:* *: :****:*****.** *** *** :** **: *** .:* **
Rv2509|M.tuberculosis_H37Rv ARLADKYRVTVDVRPADLADPQERSKLADELAARPISILCANAGTATFGP
Mb2537|M.bovis_AF2122/97 ARLADKYRVTVDVRPADLADPQERSKLADELAARPISILCANAGTATFGP
MLBr_00429|M.leprae_Br4923 ARLADKYRVAVEVRPADLTDPSERVKLADELTVRPISILCANAGTATFGP
MMAR_3856|M.marinum_M ARLTDKYRVTVEVRPADLADPQERTKLTDELAARPISILCANAGTATFGP
MUL_3788|M.ulcerans_Agy99 ARLTDKYRVSVEVRPADLADPQERTKLTDELAARPISILCANAGTATFGP
MAV_1666|M.avium_104 ARLTEKYRVTVEVRPADLADAGERATLCDELAARPISVLCANAGTATFGP
MAB_1537c|M.abscessus_ATCC_199 NALSEKTGVIVEVRAVDLADPGARDALCTELSERNISILCNNAGTATFGP
Mflv_2545|M.gilvum_PYR-GCK DRLRSEFGVTVEVRAVDLADPAARDVFCEELATRTISILCANAGTATFGA
Mvan_4098|M.vanbaalenii_PYR-1 ERLRQKYGVTVEVRAVDLADPVSRGAFCDELAGRNISILCANAGTATFGA
MSMEG_4722|M.smegmatis_MC2_155 QRLTERYGVTVEVRAVDLTDPAARATLCDELAEREISILCANAGTATFGA
TH_1383|M.thermoresistible__bu DRLTDRHGVTVEVRPLDLADPEQRGTLCAELAEREISILCANAGVATFGP
* .. * *:**. **:*. * : **: * **:** ***.****.
Rv2509|M.tuberculosis_H37Rv IASLDLAGEKTQVQLNAVAVHDLTLAVLPGMIERKAGGILISGSAAGNSP
Mb2537|M.bovis_AF2122/97 IASLDLAGEKTQVQLNAVAVHDLTLAVLPGMIERKAGGILISGSAAGNSP
MLBr_00429|M.leprae_Br4923 VSALDPAGEKAQVQLNAVAVHDLTLAVLPGMIERKAGGILISGSAAGNSP
MMAR_3856|M.marinum_M VASLDPAGEKAQVQLNAVAVHDLTLAVLPGMIERRAGGILISGSAAGNSP
MUL_3788|M.ulcerans_Agy99 VASLDPAGEKAQVQLNAVAVHDLTLAVLPGMIERRAGGILISGSAAGNSP
MAV_1666|M.avium_104 VATLDPAGEKAQVQLNVLGVHDLTLAVLPGMVERRAGGILISGSAAGNSP
MAB_1537c|M.abscessus_ATCC_199 IRGLDPAGERKQVQLNAVAVHDLTLAVLPGMLARGAGGILISGSAAGNSP
Mflv_2545|M.gilvum_PYR-GCK VASLDPADERKQVQLNVLGVHDLVLAVLPGMVARRAGGILISGSAAGNSP
Mvan_4098|M.vanbaalenii_PYR-1 VHALDPAEERKQVQLNVLGVHDLVLAVLPGMVQRRAGGILISGSAAGNSP
MSMEG_4722|M.smegmatis_MC2_155 VKDLDPAGEKAQVQLNVLGVHDLVLAVLPGMVARRAGGILISGSAAGNSP
TH_1383|M.thermoresistible__bu VHKLDPAEEKAQVQLNVLGVHDLVLAVLPGMVQRRAGGILISGSAAGNSP
: ** * *: *****.:.****.*******: * ***************
Rv2509|M.tuberculosis_H37Rv IPYNATYAATKAFVNTFSESLRGELRGSGVHVTVLAPGPVRTELPDASEA
Mb2537|M.bovis_AF2122/97 IPYNATYAATKAFVNTFSESLRGELRGSGVHVTVLAPGPVRTELPDASEA
MLBr_00429|M.leprae_Br4923 IPYNATYAATKAFANTFSESLRGELHGSGVHVTLLAPGPVRTELPDHDEA
MMAR_3856|M.marinum_M IPYNATYAATKAFANTFSESLRGELRGSGVHVTLLAPGPVRTDLPDASEA
MUL_3788|M.ulcerans_Agy99 IPYNATYAATKAFANTFSESLRGELRGSGVHVTLLAPGPVRTDLPDASEA
MAV_1666|M.avium_104 IPYNATYAATKAFVNTFSESLRGELRGSGVHVTLLAPGPVRTDLPDDAEA
MAB_1537c|M.abscessus_ATCC_199 IPNNATYAATKAFANTFSESLRGELKGTGVHVTLLAPGPVRTVEPDPAEA
Mflv_2545|M.gilvum_PYR-GCK IPNNATYAATKAFANTFGESLRGEVKSTGVHVTVLAPGPVRTELPDPDEQ
Mvan_4098|M.vanbaalenii_PYR-1 IPNNATYAATKAFANTFGESLRGEVKSAGVHVTVLAPGPVRTELPDPDEQ
MSMEG_4722|M.smegmatis_MC2_155 IPNNATYAASKAFANTFSESLRGELTGVGVHVTLLAPGPVRTELPDPSEQ
TH_1383|M.thermoresistible__bu IPNNATYAATKAFANTFSESLRGELKDLGVHVTLLAPGPVRSDLPDPSEA
** ******:***.***.******: . *****:*******: ** *
Rv2509|M.tuberculosis_H37Rv SLVEKLVPDFLWISTEHTARVSLNALERNKMRVVPGLTSKAMSVASQYAP
Mb2537|M.bovis_AF2122/97 SLVEKLVPDFLWISTEHTARVSLNALERNKMRVVPGLTSKAMSVASQYAP
MLBr_00429|M.leprae_Br4923 SLVEKLVPDFLWISTEHTARLSLDALERNKMRVVPGLTSKAMSVASRYAP
MMAR_3856|M.marinum_M SLVERLVPDFLWISTEHTAQVSLDALARNKMRVVPGLTSKAMSVASQYAP
MUL_3788|M.ulcerans_Agy99 SLVERLVPDFLWISTEHTAQVSLDALARNKMRVVAGLTSKAMSVASQYAP
MAV_1666|M.avium_104 SIVERLVPDFLWISTEHTARVSLDALGRNKMRVVPGLTSKAMSVASGYAP
MAB_1537c|M.abscessus_ATCC_199 SIVDKVVPDFLWINGEYTARVSLNALERNKMRVVPGITSKAMSVAAGYAP
Mflv_2545|M.gilvum_PYR-GCK SLVERLIPDFLWIDTEYTAKVSLDGLDRNKMRVVPGVTSKAMSAASGYAP
Mvan_4098|M.vanbaalenii_PYR-1 SLVERLIPDFLWIDTEYTAKVSLDGLDRNKMRVVPGITSKAMSAASGYAP
MSMEG_4722|M.smegmatis_MC2_155 SLVERLIPDFLWIDTEYTAKLSLDGLEHNKMRVVPGVTSKAMSVASGYAP
TH_1383|M.thermoresistible__bu SLVDRLVPDFLWISTEHTAKLSLDGLAANKMRVVPGLTSKAMSVASGYAP
*:*::::******. *:**::**:.* ******.*:******.*: ***
Rv2509|M.tuberculosis_H37Rv RAIVAPIVGAFYKRLGGS
Mb2537|M.bovis_AF2122/97 RAIVAPIVGAFYKRLGGS
MLBr_00429|M.leprae_Br4923 RAIVASIVGNFYKKLGGG
MMAR_3856|M.marinum_M RAIVAPIVGGFYKKLGGG
MUL_3788|M.ulcerans_Agy99 RAIVAPIVGGFYKKLGGG
MAV_1666|M.avium_104 RAIVAPIVGSFYKKLGGG
MAB_1537c|M.abscessus_ATCC_199 RAVVAPIVGSFYKKLGD-
Mflv_2545|M.gilvum_PYR-GCK RAIVAPIVGAVYKKLGGD
Mvan_4098|M.vanbaalenii_PYR-1 RAIVAPIVGAVYKKLGGD
MSMEG_4722|M.smegmatis_MC2_155 RAIVTPIVGAVYKKLGGG
TH_1383|M.thermoresistible__bu RAIVAPIVGAFYKKLGGG
**:*:.*** .**:**.