For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGARRATYWAVLDTLVVGYALLPVLWIFSLSLKPTSTVKDGKLIPSTVTFDNYRGIFRGDLFSSALINSI GIGLITTVIAVVLGAMAAYAVARLEFPGKRLLIGAALLITMFPSISLVTPLFNIERAIGLFDTWPGLILP YITFALPLAIYTLSAFFREIPWDLEKAAKMDGATPGQAFRKVIVPLAAPGLVTAAILVFIFAWNDLLLAL SLTATKAAITAPVAIANFTGSSQFEEPTGSIAAGAIVITIPIIVFVLIFQRRIVAGLTSGAVKG
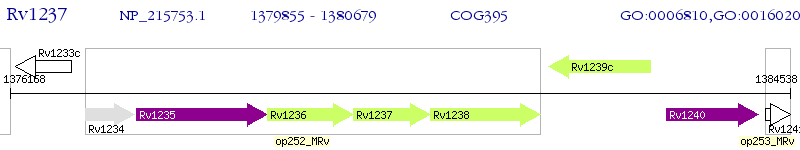
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1237 | sugB | - | 100% (274) | PROBABLE SUGAR-TRANSPORT INTEGRAL MEMBRANE PROTEIN ABC TRANSPORTER SUGB |
| M. tuberculosis H37Rv | Rv2039c | - | 1e-28 | 28.63% (255) | sugar-transport integral membrane protein ABC transporter |
| M. tuberculosis H37Rv | Rv2317 | uspB | 3e-24 | 30.90% (233) | sugar-transport integral membrane protein ABC transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1269 | sugB | 1e-151 | 100.00% (274) | sugar-transport integral membrane protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_3853 | - | 1e-91 | 57.51% (273) | binding-protein-dependent transport systems inner membrane |
| M. leprae Br4923 | MLBr_01088 | - | 1e-137 | 89.78% (274) | putative ABC-transport protein, inner membrane component |
| M. abscessus ATCC 19977 | MAB_1374 | - | 1e-125 | 81.48% (270) | sugar ABC transporter, permease protein SugB |
| M. marinum M | MMAR_4204 | sugB | 1e-137 | 90.11% (273) | sugar-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_1376 | sugB | 1e-133 | 89.39% (264) | ABC transporter, permease protein SugB |
| M. smegmatis MC2 155 | MSMEG_5059 | - | 1e-131 | 85.04% (274) | ABC transporter, permease protein SugB |
| M. thermoresistible (build 8) | TH_2867 | - | 1e-131 | 86.28% (277) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4511 | sugB | 1e-121 | 90.24% (246) | sugar-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2542 | - | 4e-91 | 57.51% (273) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1237|M.tuberculosis_H37Rv -------------MGARRATYWAVLDTLVVGYALLPVLWIFSLSLKPTST
Mb1269|M.bovis_AF2122/97 -------------MGARRATYWAVLDTLVVGYALLPVLWIFSLSLKPTST
MLBr_01088|M.leprae_Br4923 MGIEKVKVTAGVRVGARRATIWVIIDTFVVGYALLPVLWILSLSLKPAST
MMAR_4204|M.marinum_M -----------MTTGSGRRTFWVVIDTMVVVYALLPVLWILSLSLKPTST
MUL_4511|M.ulcerans_Agy99 ----------------------------------------MSLSPKPTST
MAV_1376|M.avium_104 -----------------------MIDTLVVVYALLPVLWIFSLSLKPTST
MSMEG_5059|M.smegmatis_MC2_155 ---------MADRVDARRATWWSVVNILVIVYALIPVLWILSLSLKPTSS
TH_2867|M.thermoresistible__bu ---------VNERVTARRATGWIVIDLLVILYALFPVLWILSLSLKSTAT
MAB_1374|M.abscessus_ATCC_1997 ----------MSRPDSSRTAGWLIADVLVLCYALVPVLWVLSLSLKPTSS
Mflv_3853|M.gilvum_PYR-GCK --------------MTKNTKWWTIAGIVIVIYSFVPMLWMISLAFKPPSD
Mvan_2542|M.vanbaalenii_PYR-1 --------------MTKNTKWWTIAGIVIVIYSFVPMLWMISLAFKPPSD
:**: *..:
Rv1237|M.tuberculosis_H37Rv VKDGK--LIPSTVTFDNYRGIFRG---DLFSSALINSIGIGLITTVIAVV
Mb1269|M.bovis_AF2122/97 VKDGK--LIPSTVTFDNYRGIFRG---DLFSSALINSIGIGLITTVIAVV
MLBr_01088|M.leprae_Br4923 VKDGK--LIPSLVTFDNYRGIFRS---DLFSSALINSIGIGLTTTVIAVM
MMAR_4204|M.marinum_M VKDGR--LIPSSVTLDNYRGVFGG---DLFSSALINSIGIGLTTTVIAVL
MUL_4511|M.ulcerans_Agy99 VKDGR--LIPSSVTLDNYRGVFGG---DLFSSALINSIGIGLTTTVIAVL
MAV_1376|M.avium_104 VKDGK--LIPSAISLENYRGIFRG---DFFSSALINSVGIGLITTAVAVL
MSMEG_5059|M.smegmatis_MC2_155 VKDGK--LIPTEITFANYKAIFSG---DAFTSALFNSIGIGLITTIIAVV
TH_2867|M.thermoresistible__bu VKDGK--LIPDEITFDNYKAIFRGGGSGVFTSALINSIGIGLITTVIAVV
MAB_1374|M.abscessus_ATCC_1997 VKDGK--FFPWPITLDNYRGIFSG---NVFTSALVNSIGIGLIATVIAVS
Mflv_3853|M.gilvum_PYR-GCK IVSGEPSFLPSTVTFDNFAQIFSN---PLFTRALINSIGIALVATVISVI
Mvan_2542|M.vanbaalenii_PYR-1 IVSGKPSFLPSSITFDNFAQIFSQ---PLFTRALINSIGIAVIATVISVI
: .*. ::* ::: *: :* *: **.**:**.: :* ::*
Rv1237|M.tuberculosis_H37Rv LGAMAAYAVARLEFPGKRLLIGAALLITMFPSISLVTPLFNIERAIGLFD
Mb1269|M.bovis_AF2122/97 LGAMAAYAVARLEFPGKRLLIGAALLITMFPSISLVTPLFNIERAIGLFD
MLBr_01088|M.leprae_Br4923 FGAMAAYAIARLAFPGKRLLVGTTLLVTMFPAISLVTPLFNIERRVGLFD
MMAR_4204|M.marinum_M LGAMAAYAVARLDFPGKRLLVGVALLITMFPAISLVTPLFNIERRIGLFD
MUL_4511|M.ulcerans_Agy99 LGAMAACAVARLDFPGKRLLVGVTLLITMFPAISLVTPLFNIERRIGLFD
MAV_1376|M.avium_104 LGAMAAYAVARLDFPGKRLLIGATLLITMFPAISLVTPLFNIERFLGLFD
MSMEG_5059|M.smegmatis_MC2_155 IGGMAAYAVARLQFPGKQLLIGVALLIAMFPHISLVTPIFNMWRGIGLFD
TH_2867|M.thermoresistible__bu VGGMAAYAVARLNFPGKRLLIGVALLIAMFPQISLVTPLFNIERQLGLFD
MAB_1374|M.abscessus_ATCC_1997 VGTMAAYAVARLDFPGKKALIGAALLIAMFPQISLVTPIFNIERSVGLFD
Mflv_3853|M.gilvum_PYR-GCK IAMFAAYAIARLEFPGKKVLLSLALGIAMFPQAALVGPLFDMWRGLGIYD
Mvan_2542|M.vanbaalenii_PYR-1 IAMFAAYAIARLEFPGKKVLLSLALGIAMFPQAALVGPLFDMWRGLGIYD
.. :** *:*** ****: *:. :* ::*** :** *:*:: * :*::*
Rv1237|M.tuberculosis_H37Rv TWPGLILPYITFALPLAIYTLSAFFREIPWDLEKAAKMDGATPGQAFRKV
Mb1269|M.bovis_AF2122/97 TWPGLILPYITFALPLAIYTLSAFFREIPWDLEKAAKMDGATPGQAFRKV
MLBr_01088|M.leprae_Br4923 TWAGLILPYITFALPLAIYTLSAFFAEIPWDLEKAAKMDGATSGQAFRKV
MMAR_4204|M.marinum_M TWPGLILPYITFALPLAIYTLSAFFREIPWDLEKAAQMDGATPYQAFRKV
MUL_4511|M.ulcerans_Agy99 TWPELILPYITFALPLAIYTLSAFFREIPWDLEKAAQMDGATPYQAFRKV
MAV_1376|M.avium_104 TWPGLILPYITFALPLAIYTLSAFFREIPWDLEKAAKIDGATPAQAFRKV
MSMEG_5059|M.smegmatis_MC2_155 TWPGLIIPYITFALPLAIYTLSAFFREIPWDLEKAAKMDGATPAQAFRKV
TH_2867|M.thermoresistible__bu TWAGLIIPYITFALPLAIYTLSAFFREIPWDLEKAAKMDGATPGQAFRKV
MAB_1374|M.abscessus_ATCC_1997 TWPGLIIPYITFALPLAIYTLSAFFREIPWELEKAAKMDGATPAQAFWKV
Mflv_3853|M.gilvum_PYR-GCK TWLGLIIPYLTFALPLSIWTMSAFFRQIPWEMEHAAQVDGATQWQAFRKV
Mvan_2542|M.vanbaalenii_PYR-1 TWLGLIIPYLTFALPLSIWTMSAFFRQIPWEMEHAAQVDGATQWQAFRKV
** **:**:******:*:*:**** :***::*:**::**** *** **
Rv1237|M.tuberculosis_H37Rv IVPLAAPGLVTAAILVFIFAWNDLLLALSLTATKAAITAPVAIANFTGSS
Mb1269|M.bovis_AF2122/97 IVPLAAPGLVTAAILVFIFAWNDLLLALSLTATKAAITAPVAIANFTGSS
MLBr_01088|M.leprae_Br4923 IVPLAAPGLVTAAILVFIFAWNDLLLALSLTSTKAAITAPVAITNFTGSS
MMAR_4204|M.marinum_M IVPLAAPGLVTAAILVFIFAWNDLLLALSLTATKASITAPVAIANFAGSS
MUL_4511|M.ulcerans_Agy99 IVPLAAPGLVTAAILVFIFAWNDLLLALSLTATKASITAPIAIANFAGSS
MAV_1376|M.avium_104 IVPLAAPGVVTAAILVFIFAWNDLLLALTLTATKAAITAPVAIVSFSGSS
MSMEG_5059|M.smegmatis_MC2_155 IAPLAAPGIVTAAILVFIFAWNDLLLALSLTATQRAITAPVAIANFTGSS
TH_2867|M.thermoresistible__bu IAPLAAPGIVTAAILVFIFAWNDLLLALSLTATERAITAPVAIANFTGSS
MAB_1374|M.abscessus_ATCC_1997 IAPLATPGIVTSAILVFIFAWNDLLLAISLTATDRSITAPVAIANFTGSS
Mflv_3853|M.gilvum_PYR-GCK IVPLAAPGVFTTAILTFFFCWNDFLFAISLTSTDSARTVPAALAFFQGAS
Mvan_2542|M.vanbaalenii_PYR-1 IVPLAAPGVFTTAILTFFFCWNDFLFAISLTSTDSARTVPAALAFFQGAS
*.***:**:.*:***.*:*.***:*:*::**:*. : *.* *:. * *:*
Rv1237|M.tuberculosis_H37Rv QFEEPTGSIAAGAIVITIPIIVFVLIFQRRIVAGLTSGAVKG
Mb1269|M.bovis_AF2122/97 QFEEPTGSIAAGAIVITIPIIVFVLIFQRRIVAGLTSGAVKG
MLBr_01088|M.leprae_Br4923 QFEEPTGSIAAGAIVITAPIIVFVWIFQRRIVAGLTSGAVKG
MMAR_4204|M.marinum_M QFEEPTGSIAAGAMVITVPIIVFVLIFQRRIVAGLTSGAVKG
MUL_4511|M.ulcerans_Agy99 QFEEPTGSIAAGAMVITVPIIVFVLIFQLRIVAGLTSGAVKG
MAV_1376|M.avium_104 QFEEPTGSIAAGAVVITVPVILFVLIFQRRIVAGLTSGAVKG
MSMEG_5059|M.smegmatis_MC2_155 QFEEPTGSIAAGAMVITIPIIIFVLIFQRRIVAGLTSGAVKG
TH_2867|M.thermoresistible__bu QFEEPTGSIAAGAMVITIPIIVFVLIFQRRIVSGLTSGAVKG
MAB_1374|M.abscessus_ATCC_1997 QFEEPTGSIAAGAIVITVPIIIFVLIFQRRIVAGLTSGAVKG
Mflv_3853|M.gilvum_PYR-GCK YFESPVPYIMAASVIVTIPVVILVLVFQRRIVAGLTSGAVKG
Mvan_2542|M.vanbaalenii_PYR-1 YFESPVPYIMAASVIVTIPVVILVLIFQRRIVAGLTSGAVKG
**.*. * *.::::* *::::* :** ***:*********