For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTGGPLAGVKVIELGGIGPGPHAGMVLADLGADVVRVRRPGGLTMPSEDRDLLHRGKRIVDLDVKTQPQ AMLELAAKADVLLDCFRPGTCERLGIGPDDCASVNPRLIFARITGWGQDGPLASTAGHDINYLSQTGALA AFGYADRPPMPPLNLVADFGGGSMLVLLGIVVALYERERSGVGQVVDAAMVDGVSVLAQMMWTMKGIGSL RDQRESFLLDGGAPFYRCYETSDGKYMAVGAIEPQFFAALLSGLGLSAADVPTQLDVAGYPQMYDIFAER FASRTRDEWTRVFAGTDACVTPVLAWSEAANNDHLKARSTVITAHGVQQAAPAPRFSRTPAGPVRPPPAA ATPIDEINW
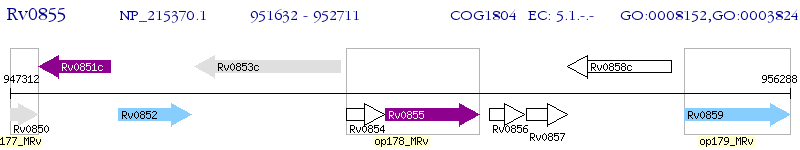
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0855 | far | - | 100% (359) | PROBABLE FATTY-ACID-CoA RACEMASE FAR |
| M. tuberculosis H37Rv | Rv1143 | mcr | e-113 | 57.64% (347) | alpha-methylacyl-CoA racemase |
| M. tuberculosis H37Rv | Rv3272 | - | 1e-30 | 40.10% (202) | hypothetical protein Rv3272 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0878 | far | 0.0 | 100.00% (359) | fatty-acid-CoA racemase |
| M. gilvum PYR-GCK | Mflv_2126 | - | 1e-106 | 54.47% (347) | L-carnitine dehydratase/bile acid-inducible protein F |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0845 | - | 1e-148 | 69.01% (355) | fatty-acid-CoA racemase Far |
| M. marinum M | MMAR_4681 | far | 0.0 | 88.86% (359) | fatty-acid-CoA racemase Far |
| M. avium 104 | MAV_0976 | - | 1e-179 | 83.84% (359) | alpha-methylacyl-CoA racemase |
| M. smegmatis MC2 155 | MSMEG_5734 | - | 1e-162 | 74.93% (359) | alpha-methylacyl-CoA racemase |
| M. thermoresistible (build 8) | TH_4477 | - | 1e-108 | 54.14% (362) | PUTATIVE L-carnitine dehydratase/bile acid-inducible protein F |
| M. ulcerans Agy99 | MUL_0986 | mcr | 1e-109 | 54.75% (358) | alpha-methylacyl-CoA racemase Mcr |
| M. vanbaalenii PYR-1 | Mvan_4578 | - | 1e-113 | 56.78% (354) | L-carnitine dehydratase/bile acid-inducible protein F |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0855|M.tuberculosis_H37Rv MTTG----GPLAGVKVIELGGIGPGPHAGMVLADLGADVVRVRRPG---G
Mb0878|M.bovis_AF2122/97 MTTG----GPLAGVKVIELGGIGPGPHAGMVLADLGADVVRVRRPG---G
MMAR_4681|M.marinum_M MAGG----GPLAGVKVIELGGIGPGPHAGMVLADLGADVVRVRRPG---G
MAV_0976|M.avium_104 MSSG----GPLAGVRVIELGGIGPGPHAGMVLADLGADVVRVRRPG---V
MSMEG_5734|M.smegmatis_MC2_155 MTKT----GPLSGVRVVELGGIGPGPHAAMMLSDLGADVVRVRRPG---G
MAB_0845|M.abscessus_ATCC_1997 MADTGVKTGPLAGVKVIELGGIGPGPHAAMMLSDLGADVIRVRRPG---G
Mflv_2126|M.gilvum_PYR-GCK MAG------PLQGLRVVELAGIGPGPHAAMIFGDLGADVVRIERPGKGPG
Mvan_4578|M.vanbaalenii_PYR-1 MAG------PLQGLRVVELAGIGPGPHAAMILGDLGADVVRIERPGKGPG
TH_4477|M.thermoresistible__bu MAG------PLQGLRVVELAGIGPGPHAAMILGDLGADVVRIERPRNTPG
MUL_0986|M.ulcerans_Agy99 MSG------PLNGLRVVELAGIGPGPHAAMILGDLGADVVRIDRPSSSAG
*: ** *::*:**.********.*::.******:*: **
Rv0855|M.tuberculosis_H37Rv LTMPSEDRDLLHRGKRIVDLDVKTQP--QAMLELAAKADVLLDCFRPGTC
Mb0878|M.bovis_AF2122/97 LTMPSEDRDLLHRGKRIVDLDVKTQP--QAMLELAAKADVLLDCFRPGTC
MMAR_4681|M.marinum_M LQMPAEDRDLLHRGKRIVDLDVKSQP--QALLELAAKADVLLDCFRPGTC
MAV_0976|M.avium_104 AAMPSEDRDLLHRGKRIVDLDVKARP--QALLDLAAKADVLLDCFRPGTC
MSMEG_5734|M.smegmatis_MC2_155 LTLPAENVDLLHRGKRIVDLDVKAEP--AKLLDLVAKADVLLDCFRPGTC
MAB_0845|M.abscessus_ATCC_1997 LAIPAEETDLFHRGKRLVNLDVKKDP--EALLALVDKADVLLDPFRPGVC
Mflv_2126|M.gilvum_PYR-GCK PATKPGG-DYLLRNRRSVAANLKSDEDREMVLRLIAKADVLIEGFRPGVT
Mvan_4578|M.vanbaalenii_PYR-1 PATKPGG-DYLLRNRRSVAANLKSEEDRDMVLRLIAKADVLIEGFRPGVT
TH_4477|M.thermoresistible__bu PGIPRGAKDHLLRNRRSVAADLKTEEGKELVRKLIAKADVLIEGYRPGVT
MUL_0986|M.ulcerans_Agy99 -GRGGGVKDAMLRNRRIVTADLKSDQGRELVLKLVAKADVLIEGYRPGVT
* : *.:* * ::* : * *****:: :***.
Rv0855|M.tuberculosis_H37Rv ERLGIGPDDCASVNPRLIFARITGWGQDGPLASTAGHDINYLSQTGALAA
Mb0878|M.bovis_AF2122/97 ERLGIGPDDCASVNPRLIFARITGWGQDGPLASTAGHDINYLSQTGALAA
MMAR_4681|M.marinum_M ERLGIGPDDCAAVNPRLIFARITGWGQDGPLAPTAGHDINYLSQTGALSA
MAV_0976|M.avium_104 ERLGIGPDDCAAVNPRLIFARITGWGQHGPLAQTAGHDINYLSQTGVLSA
MSMEG_5734|M.smegmatis_MC2_155 ERLGIGPKECEAVNPRLIFARITGWGQDGPLAQTAGHDINYLSQTGALSA
MAB_0845|M.abscessus_ATCC_1997 ERIGIGPEECAKRNPRLIFARMTGWGQHGPMADRAGHDINYLSLTGALGS
Mflv_2126|M.gilvum_PYR-GCK ERLGLGPEDCAKVNEKLVYARMTGWGQDGPRAQQAGHDINYISLNGVLHA
Mvan_4578|M.vanbaalenii_PYR-1 ERLGLGPHDCAKVNEKLIYARMTGWGQDGPRAQQAGHDINYISLNGALHA
TH_4477|M.thermoresistible__bu ERLGLGPEDCAKINPRLIYGRMTGWGQDGPRAQQAGHDINYISLNGTLHA
MUL_0986|M.ulcerans_Agy99 ERLGLGPQDCAKVNDRLIYARMTGWGQAGPRSQQAGHDINYISLNGILHA
**:*:**.:* * :*::.*:***** ** : *******:* .* * :
Rv0855|M.tuberculosis_H37Rv FGYADRPPMPPLNLVADFGGGSMLVLLGIVVALYERERSGVGQVVDAAMV
Mb0878|M.bovis_AF2122/97 FGYADRPPMPPLNLVADFGGGSMLVLLGIVVALYERERSGVGQVVDAAMV
MMAR_4681|M.marinum_M LGYRDRPPTPPLNLVADFGGGSMLVLLGIMAALYERERSGQGQVIDAAMV
MAV_0976|M.avium_104 LGHADRPPLPPLNLVADFGGGSMLVLLGITAALYERERSGRGQVIDAAMV
MSMEG_5734|M.smegmatis_MC2_155 IGYRDRPPVAPMNLVADFGGGSMLVLVGIVSALYERERSGKGQVIDAAMV
MAB_0845|M.abscessus_ATCC_1997 MGYKDRPPMPPMNLVADFGGGSMLLVQGILAALYEREKSGKGQVIDGAMV
Mflv_2126|M.gilvum_PYR-GCK IGRHGERPVPPLNLAGDFGGGSMFLLVGVLAALWERERSGKGQVVDAAMV
Mvan_4578|M.vanbaalenii_PYR-1 IGRAGERPVPPLNLVGDFGGGSMFLLVGVLSALYERERSGKGQVVDAAMV
TH_4477|M.thermoresistible__bu VGRKGERPVPPLNLAGDFGGGSMFLLVGILAALFERQTSGKGQVVDAAMI
MUL_0986|M.ulcerans_Agy99 IGRVGERPVPPLNLVGDFGGGSMFLLVGILAALWERHNSGKGQVIDAAMV
.* .. * .*:**..*******::: *: **:**. ** ***:*.**:
Rv0855|M.tuberculosis_H37Rv DGVSVLAQMMWTMKGIGSLRDQRESFLLDGGAPFYRCYETSDGKYMAVGA
Mb0878|M.bovis_AF2122/97 DGVSVLAQMMWTMKGIGSLRDQRESFLLDGGAPFYRCYETSDGKYMAVGA
MMAR_4681|M.marinum_M DGVSVLAQMMWTMKGIGSLRDERESFLLDGGAPFYRCYETSDGKHMAVGA
MAV_0976|M.avium_104 DGVSLLAQMMWTMKSTGALRDRRESFLLDGGAPFYGCYETADGKYVAVGA
MSMEG_5734|M.smegmatis_MC2_155 DGVSMLAQMMWTMRATGSLRDERESFLLDGGAPFYRCYETADGGYMAVGS
MAB_0845|M.abscessus_ATCC_1997 DGVSQLAQMQWTMYNNGLLFDEREAGLLDGGSPFYGTYETSDGKYMAVGS
Mflv_2126|M.gilvum_PYR-GCK DGSSVLSMMMWSFRGMGMWSDERGVNMLDTGAPYYDTYECSDGRHIAVGC
Mvan_4578|M.vanbaalenii_PYR-1 DGSSVLSMMMWAFRGMGMWSDERGTNMLDTGAPYYDTYETADGKYVAVGA
TH_4477|M.thermoresistible__bu DGSAVLMQMMWAFRAMGMWSDERGVNMLDTGAPYYDTYETADGKYVAVGA
MUL_0986|M.ulcerans_Agy99 DGSSVLVQMMWAMRGIGIWSDVRGTNMLDGGAPYYDTYECADGGYVAVGA
** : * * *:: * * * :** *:*:* ** :** ::***.
Rv0855|M.tuberculosis_H37Rv IEPQFFAALLSGLGLSAADVPTQLDVAGYPQMYDIFAERFASRTRDEWTR
Mb0878|M.bovis_AF2122/97 IEPQFFAALLSGLGLSAADVPTQLDVAGYPQMYDIFAERFASRTRDEWTR
MMAR_4681|M.marinum_M IEPQFFAALLAGLGLSAAEVPSQLDIAAYPKMREMFSERFASRTRDEWAA
MAV_0976|M.avium_104 IEPQFFAALLDGLGLSPEEVPGQLDRDSYPRMREIFTQRFAGRTRDEWAR
MSMEG_5734|M.smegmatis_MC2_155 IEPQFFAQLVAGLGLAADEIPGQFEIGRYDEMRKIFTERFKTKTRAEWTE
MAB_0845|M.abscessus_ATCC_1997 IEPQFFAILVQGLGLDPESVPFQLDKARYPEMRKMFDDAFKTKTRDEWTE
Mflv_2126|M.gilvum_PYR-GCK IEPQFYAEFLKGVGLEGADLPGQNDMGRWPELREAFSKAIAAHDRDHWAG
Mvan_4578|M.vanbaalenii_PYR-1 IEPQFYAEMLKGIGLDGADLPDQNDMSRWPELREAFTKAFAAHDRDHWAK
TH_4477|M.thermoresistible__bu IEPQFYAEMLKGLGLEGEDLPDQNDMSRWDELRARFAEVFKSKDRDHWAQ
MUL_0986|M.ulcerans_Agy99 IEPQFYAAMLAGLGLDAVNLPPQNDVARWPELRAVLTEAFGRYDRDHWAK
*****:* :: *:** .:* * : : .: : . : * .*:
Rv0855|M.tuberculosis_H37Rv VFAGTDACVTPVLAWSEAANNDHLKARSTVIT-AHGVQQAAPAPRFSRTP
Mb0878|M.bovis_AF2122/97 VFAGTDACVTPVLAWSEAANNDHLKARSTVIT-AHGVQQAAPAPRFSRTP
MMAR_4681|M.marinum_M VFAGTDACVTPVLKWSEAAASDHLRARSTVIS-AHGVEQAAPAPRFSRTP
MAV_0976|M.avium_104 VFAGTDACVTPVLTWSEAAASEHLRARSTVIT-AHGVDQAAPAPRFSHTP
MSMEG_5734|M.smegmatis_MC2_155 IFAGTDACVTPVLTWGEAAENAHLRERSTLID-VDGVAQAAPAPRFSRTP
MAB_0845|M.abscessus_ATCC_1997 IFIGTDACVSPVLTWGEAKQNEHLRDRGTIVD-VDGVTQAAPAPRFSRTP
Mflv_2126|M.gilvum_PYR-GCK VFAGTDACVTPVLSFAEVESEPHNTERDTFYS-ENGSLYPAPAPRFSRSV
Mvan_4578|M.vanbaalenii_PYR-1 VFAGTDACVTPVLSFAEVESEPHNTERDTFYS-ENGSLYPAPAPRFSRSA
TH_4477|M.thermoresistible__bu VFAGTDACVTPVLSFAEVESEAHVVERNTFYR-EGDHLLPAPAPRFSRSV
MUL_0986|M.ulcerans_Agy99 VFADSDACVTPVLSFGEVHTEPHIAERNTFYEGDDGGPQPMPAPRFSRTV
:* .:****:*** :.*. . * *.*. . . ******::
Rv0855|M.tuberculosis_H37Rv AGPVRPP--PAAATPIDEINW-
Mb0878|M.bovis_AF2122/97 AGPVRPP--PAAATPIDEINW-
MMAR_4681|M.marinum_M AGPVGPP--PATITPIGEINW-
MAV_0976|M.avium_104 ARPVGRP--PATSTPLDEINW-
MSMEG_5734|M.smegmatis_MC2_155 APTPGKP--PQGSTSIEDIGWD
MAB_0845|M.abscessus_ATCC_1997 SGPLGAP--PKDTTELSDIGW-
Mflv_2126|M.gilvum_PYR-GCK PSAPKAPGVPGADTEAVLQEWV
Mvan_4578|M.vanbaalenii_PYR-1 PSAPKPPGVPGADTEAVLREWV
TH_4477|M.thermoresistible__bu PSAPTPPRDPGADTDAVLQDWQ
MUL_0986|M.ulcerans_Agy99 PSRPRPPAAPLTDVEAVLSDWA
. * * . *