For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRMAEKPISPTKTRTRFEDIQAHYDVSDDFFALFQDPTRTYSCAYFEPPELTLEEAQYAKVDLNLDKLDL KPGMTLLDIGCGWGTTMRRAVERFDVNVIGLTLSKNQHARCEQVLASIDTNRSRQVLLQGWEDFAEPVDR IVSIEAFEHFGHENYDDFFKRCFNIMPADGRMTVQSSVSYHPYEMAARGKKLSFETARFIKFIVTEIFPG GRLPSTEMMVEHGEKAGFTVPEPLSLRPHYIKTLRIWGDTLQSNKDKAIEVTSEEVYNRYMKYLRGCEHY FTDEMLDCSLVTYLKPGAAA*
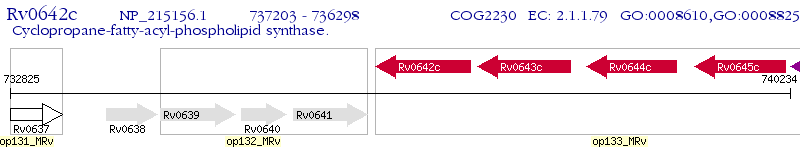
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0642c | mmaA4 | - | 100% (301) | METHOXY MYCOLIC ACID SYNTHASE 4 MMAA4 (METHYL MYCOLIC ACID SYNTHASE 4) (MMA4) (HYDROXY MYCOLIC ACID SYNTHASE) |
| M. tuberculosis H37Rv | Rv0643c | mmaA3 | e-105 | 60.41% (293) | methoxy mycolic acid synthase |
| M. tuberculosis H37Rv | Rv3392c | cmaA1 | 1e-95 | 58.74% (286) | cyclopropane-fatty-acyl-phospholipid synthase 1 CMAA1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0661c | mmaA4 | 1e-178 | 99.67% (301) | methoxy mycolic acid synthase |
| M. gilvum PYR-GCK | Mflv_5113 | - | 1e-125 | 70.45% (291) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. leprae Br4923 | MLBr_01903 | mmaA4 | 1e-159 | 87.84% (296) | methyl mycolic acid synthase 4 |
| M. abscessus ATCC 19977 | MAB_3881 | - | 2e-91 | 57.40% (277) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. marinum M | MMAR_0977 | mmaA4 | 1e-163 | 91.28% (298) | methoxy mycolic acid synthase 4, MmaA4 |
| M. avium 104 | MAV_4517 | - | 1e-160 | 88.93% (298) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. smegmatis MC2 155 | MSMEG_1351 | - | 8e-95 | 55.94% (286) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. thermoresistible (build 8) | TH_1862 | cmaA1 | 1e-126 | 72.63% (285) | CYCLOPROPANE-FATTY-ACYL-PHOSPHOLIPID SYNTHASE 1 CMAA1 |
| M. ulcerans Agy99 | MUL_0729 | mmaA4 | 1e-161 | 90.00% (300) | methoxy mycolic acid synthase 4, MmaA4 |
| M. vanbaalenii PYR-1 | Mvan_1239 | - | 1e-126 | 71.33% (293) | cyclopropane-fatty-acyl-phospholipid synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5113|M.gilvum_PYR-GCK -------------MSDNSTGTKDMTPHFEDIQAHYDLSDDFFGVFQDPTR
Mvan_1239|M.vanbaalenii_PYR-1 -------------MSDQSSGTKDMTPHFEDIQAHYDLSDDFFGVFQDPTR
TH_1862|M.thermoresistible__bu -----------------------MRPHFEDIQAHYDLSDDFFGLFQDPTR
Rv0642c|M.tuberculosis_H37Rv ----------MTRMAEKPISPTKTRTRFEDIQAHYDVSDDFFALFQDPTR
Mb0661c|M.bovis_AF2122/97 ----------MTRMAEKPISPTKTRTRFEDIQAHYDVSDDFFALFQDPTR
MMAR_0977|M.marinum_M -------------MAEIPPTPTKTRTRSEDIQAHYDVSDDFFALFQDPTR
MUL_0729|M.ulcerans_Agy99 MRRSCEHQENRTTMAEIPPTPTKMRTRSEDIQAHYDVSDDFFALFQDPTR
MAV_4517|M.avium_104 -------------MAEQPSSPTKTRTRSEDIQAHYDVSDDFFGLFQDPTR
MLBr_01903|M.leprae_Br4923 -------------MTEQPTGPTKTRTRSEDIQAHYDLSNEFFALFQDPTR
MAB_3881|M.abscessus_ATCC_1997 -------------MS-------KLTPKFSDVQAHYDLSDDFFALFLDPSR
MSMEG_1351|M.smegmatis_MC2_155 -------------MTKQPG---KLQPHFDEVQAHYDLSDDFFALFLDPSR
.: .::*****:*::**.:* **:*
Mflv_5113|M.gilvum_PYR-GCK KYSCAYFTGPNVTLSEAQIANVDQHLDALDLKPGMTLLEVGCGWGLTLQR
Mvan_1239|M.vanbaalenii_PYR-1 KYSCAYFTGPDVTLSEAQIANVDQHLDKLDLKPGMTLLEVGCGWGLTLQR
TH_1862|M.thermoresistible__bu KYSCGYFTGPDATLSDAQIADVDLNLDKLDLKPGMTLLDIGCGWGLTLRR
Rv0642c|M.tuberculosis_H37Rv TYSCAYFEPPELTLEEAQYAKVDLNLDKLDLKPGMTLLDIGCGWGTTMRR
Mb0661c|M.bovis_AF2122/97 TYSCAYFEPPELTLEEAQYAKVDLNLDKLDLKPGMTLLDIGCGWGTTMRR
MMAR_0977|M.marinum_M IYSCAYFEPPELTLEEAQYAKIDLNLDKLDLKPGMTLLDIGCGWGTTMKR
MUL_0729|M.ulcerans_Agy99 IYSCAYFEPPELTLEEAQYAKIDLNLDKLDLKPGMTLLDIGCGWGTTMKR
MAV_4517|M.avium_104 TYSCAYFARDDMSLEEAQYAKIDLNLDKLDLKPGMTLLDIGCGWGTTMKR
MLBr_01903|M.leprae_Br4923 TYSCAYFEPPDLTLEEAQYAKIDLNLNKLDLKPGMTLLDIGCGWGTTMRR
MAB_3881|M.abscessus_ATCC_1997 TYSCAYFEPETLTLEQAQQAKIDLALGKLNLEPGMTLLDVGCGWGSTMKR
MSMEG_1351|M.smegmatis_MC2_155 TYSCAYFERDDMTLEEAQLAKIDLSLGKLGLQPGMTLLDVGCGWGATMLR
***.** :*.:** *.:* *. *.*:******::***** *: *
Mflv_5113|M.gilvum_PYR-GCK AMEKYDVNVIGLTLSKNQKAYCDQLLGKLGSERTFDVRLEGWEQFHSPVD
Mvan_1239|M.vanbaalenii_PYR-1 AMEKYDVNVIGLTLSKNQKAYCDQLLSKLDTERTFDVRLEGWEQFHSPVD
TH_1862|M.thermoresistible__bu AIEKYDVNVIGLTLSRNQQAYCTQLLEQLDTNRSFDVRLMGWEQFHDPVD
Rv0642c|M.tuberculosis_H37Rv AVERFDVNVIGLTLSKNQHARCEQVLASIDTNRSRQVLLQGWEDFAEPVD
Mb0661c|M.bovis_AF2122/97 AVERLDVNVIGLTLSKNQHARCEQVLASIDTNRSRQVLLQGWEDFAEPVD
MMAR_0977|M.marinum_M AVERFDVNVIGLTLSKNQHARCESVLGEIDTNRSRQVRLHGWEDFNEPVD
MUL_0729|M.ulcerans_Agy99 AVERFDVNVIGLTLSKNQHARCESVLGEIDTNRSRQVRLHGWEDSNEPVD
MAV_4517|M.avium_104 AVERFDVNVIGLTLSKNQHARCEELLASLDSNRCRQVRLQNWEDFNEPVD
MLBr_01903|M.leprae_Br4923 AVEKYDVKVIGLTLSKNQHARCEQVLTALESNRSRQVRLQGWEDFTEPVD
MAB_3881|M.abscessus_ATCC_1997 ALEHYDVNVIGLTLSKNQFAYVNTLLDSAGSSRNYEVRLAGWEEFEGTVD
MSMEG_1351|M.smegmatis_MC2_155 ALERYDVNVVGLTLSRNQQAHVERRFSESSSPRTKRVLLAGWEEFDEPVD
*:*: **:*:*****:** * : : * * * .**: .**
Mflv_5113|M.gilvum_PYR-GCK RIVSIEAFEHFGFERYDDFFKMCFDVLPEDGRMTIQSSVGYHPYDLAERG
Mvan_1239|M.vanbaalenii_PYR-1 RIVSIEAFEHFGFERYDDFFKTCFDILPEDGRMTIQSSVGYHPYDLAERG
TH_1862|M.thermoresistible__bu RIVSIEAFEHFGFERYDHFFRNCYDIMPDDGRMTIQSSVGYHPDNLRARG
Rv0642c|M.tuberculosis_H37Rv RIVSIEAFEHFGHENYDDFFKRCFNIMPADGRMTVQSSVSYHPYEMAARG
Mb0661c|M.bovis_AF2122/97 RIVSIEAFEHFGHENYDDFFKRCFNIMPADGRMTVQSSVSYHPYEMAARG
MMAR_0977|M.marinum_M RIVSIEAFEHFGHENYDDFFKRCFDIMPEDGRMTVQSSVSYHPFEMAARG
MUL_0729|M.ulcerans_Agy99 RIVSIEAFEHFGHENYDDFFKRCFDIMPEDGRMTVQSSVSYHPFEMAARG
MAV_4517|M.avium_104 RIVSIEAFEHFGHENYDDFFKRTFDIMPDDGRMTVQSSVSYHPYDLAARG
MLBr_01903|M.leprae_Br4923 RIVSIEAFEHFGHENYDDFFKRCFNIMPSDGRMTVQSSVSYHPFDMAARG
MAB_3881|M.abscessus_ATCC_1997 RIVSIGAFEHFGHERYDSFFAMAHRVLPSDGKMLLHTIVSHHPDEWKRMG
MSMEG_1351|M.smegmatis_MC2_155 RIVSIGAFEHFGHERYDDFFRFAHRVLPDDGVMLLHTITGIHPKHAQELG
***** ******.*.** ** . ::* ** * ::: .. ** . *
Mflv_5113|M.gilvum_PYR-GCK KKLTFELARFVKFMITEIFPGGRLPTTQMMVEHGEKAGFTVPQPLSLRNH
Mvan_1239|M.vanbaalenii_PYR-1 KKLTFELARFIKFMITEIFPGGRLPTTQMMIEHGEKAGFVVPDALSLRNH
TH_1862|M.thermoresistible__bu KKLTFELARFIKFMITEIFPGGRIPTTEMMVEHGEKAGFVVPETMSLRNH
Rv0642c|M.tuberculosis_H37Rv KKLSFETARFIKFIVTEIFPGGRLPSTEMMVEHGEKAGFTVPEPLSLRPH
Mb0661c|M.bovis_AF2122/97 KKLSFETARFIKFIVTEIFPGGRLPSTEMMVEHGEKAGFTVPEPLSLRPH
MMAR_0977|M.marinum_M KKLTFETARFIKFIVTEIFPGGRLPSTGMMVEHGEKAGFVVPEPLSLRPH
MUL_0729|M.ulcerans_Agy99 KKLTFETARFIKFIVTEIFPGGRLPSTGMMVEHGEKAGFVVPEPLSLRPY
MAV_4517|M.avium_104 KKLSFETARFIKFIVTEIFPGGRLPSTQMMVDHGEKAGFVVPEPLSLRPH
MLBr_01903|M.leprae_Br4923 KKLSFETARFIKFIITEIFPGGRLPSTKMMVEHGEKAGFTVSEPLSLQSH
MAB_3881|M.abscessus_ATCC_1997 IPLLMSRVRFIYFIGTEIFPGGRLPSVPMVDKHAALAGFHITRHHSLQPH
MSMEG_1351|M.smegmatis_MC2_155 VPLTFEMARFIRFIVTEIFPGGRLPTVDMVIDHATRASFTLERRQSLQLH
* :. .**: *: ********:*:. *: .*. *.* : **: :
Mflv_5113|M.gilvum_PYR-GCK YIKTLGIWADRLEANKDAAIAATDEENYERYMRYLKGCQYYFIDEAIDVS
Mvan_1239|M.vanbaalenii_PYR-1 YIKTLGIWADRLEQNKDAAIAATDEENYRRYMRYLKGCQYYFIDEAIDVS
TH_1862|M.thermoresistible__bu YIKTLGMWADALERNKERAIAITDEENYHRYMRYLRGCQYYFIDECIDVS
Rv0642c|M.tuberculosis_H37Rv YIKTLRIWGDTLQSNKDKAIEVTSEEVYNRYMKYLRGCEHYFTDEMLDCS
Mb0661c|M.bovis_AF2122/97 YIKTLRIWGDTLQSNKDKAIEVTSEEVYNRYMKYLRGCEHYFTDEMLDCS
MMAR_0977|M.marinum_M YIKTLRIWGDNLEANKEKAIEVTSEEVYNRYIKYLRGCEHYFTDEMLDCS
MUL_0729|M.ulcerans_Agy99 YIKTLRIWGDNLEANKEKAIEVTSEEVYNRYIKYLRGCEHYFTDEMLDCS
MAV_4517|M.avium_104 YIKTLHIWGDTLESNRDKAIEVTSEEVYNRYIKYLRGCEHYFTEEMLDCS
MLBr_01903|M.leprae_Br4923 YVKTLRIWGDTLESNRAKAIEVTSEAVYNRYMKYLRGCAHYFSDEMLDCS
MAB_3881|M.abscessus_ATCC_1997 YARTLDCWATNLAAHEDEAISVQSQEVYDRYIKYLTGCAEGFREGWIDVV
MSMEG_1351|M.smegmatis_MC2_155 YARTLDMWAAALESHHDDAVRIQSQEVYDRYMHYLTGCANAFRVGYIDVN
* :** *. * :. *: .: * **::** ** * :*
Mflv_5113|M.gilvum_PYR-GCK LVTYLKPAVAA-
Mvan_1239|M.vanbaalenii_PYR-1 LVTYLKPGVAA-
TH_1862|M.thermoresistible__bu LVTYLKPGAVR-
Rv0642c|M.tuberculosis_H37Rv LVTYLKPGAAA-
Mb0661c|M.bovis_AF2122/97 LVTYLKPGAAA-
MMAR_0977|M.marinum_M LVTYLKPGAVSA
MUL_0729|M.ulcerans_Agy99 LVTYLKPGAVSA
MAV_4517|M.avium_104 LVTYLKPGAAA-
MLBr_01903|M.leprae_Br4923 LVTYLKPGALD-
MAB_3881|M.abscessus_ATCC_1997 QFTCEK------
MSMEG_1351|M.smegmatis_MC2_155 QFTLAK------
.* *