For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRGRKPRPGRIVFVGSGPGDPGLLTTRAAAVLANAALVFTDPDVPEPVVALIGTDLPPVSGPAPAEPVA GNGDAAGGGSAQEHGRAASAVVSGGPDIRPALGDPADVAKTLTAEARSGVDVVRLVAGDPLTVDAVISEV NAVARTHLHIEIVPGLAASSAVPTYAGLPLGSSHTVADVRIDPENTDWDALAAAPGPLILQATASHLAES ARSLIDHQLAESTPCVVTAHGTTCQQRSVETTLQGLTDPAVLGATDPACSANGRDSQAGPLIVTIGKTVT SRAKLNWWESRALYGWTVLVPRTKDQAGEMSERLTSYGALPVEVPTIAVEPPRSPAQMERAVKGLVDGRF QWIVFTSTNAVRAVWEKFGEFGLDARAFSGVKIACVGESTADRVRAFGISPELVPSGEQSSLGLLDDFPP YDSVFDPVNRVLLPRADIATETLAEGLRERGWEIEDVTAYRTVRAAPPPATTREMIKTGGFDAVCFTSSS TVRNLVGIAGKPHARTIIACIGPKTAETAAEFGLRVDVQPDTAAIGPLVDALAEHAARLRAEGALPPPRK KSRRR
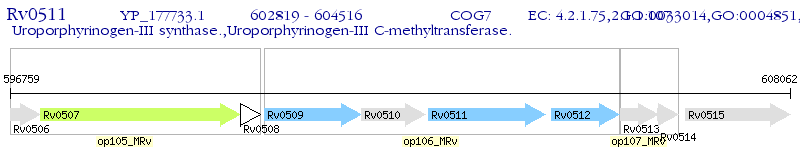
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0511 | hemD | - | 100% (565) | PROBABLE UROPORPHYRIN-III C-METHYLTRANSFERASE HEMD (UROPORPHYRINOGEN III METHYLASE) (UROGEN III METHYLASE) (SUMT) (UROGEN III METHYLASE) (UROM) |
| M. tuberculosis H37Rv | Rv2847c | cysG | 8e-19 | 29.50% (261) | multifunctional uroporphyrinogen III methylase/precorrin-2 |
| M. tuberculosis H37Rv | Rv2071c | cobM | 4e-08 | 25.10% (243) | precorrin-4 C11-methyltransferase CobM |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0524 | hemD | 0.0 | 100.00% (565) | uroporphyrin-III C-methyltransferase HemD |
| M. gilvum PYR-GCK | Mflv_0065 | - | 0.0 | 79.13% (575) | uroporphyrinogen III synthase HEM4 |
| M. leprae Br4923 | MLBr_02420 | hemD | 0.0 | 85.66% (565) | putative uroporphyrin-III C-methyltransferase |
| M. abscessus ATCC 19977 | MAB_3991c | - | 0.0 | 75.13% (567) | uroporphyrin-III C-methyltransferase |
| M. marinum M | MMAR_0844 | hemD | 0.0 | 87.99% (566) | uroporphyrin-III C-methyltransferase HemD |
| M. avium 104 | MAV_4638 | hemD | 0.0 | 88.93% (551) | uroporphyrinogen-III synthase |
| M. smegmatis MC2 155 | MSMEG_0954 | hemD | 0.0 | 78.63% (571) | uroporphyrinogen-III synthase |
| M. thermoresistible (build 8) | TH_0267 | cysG | 1e-175 | 70.50% (444) | PROBABLE UROPORPHYRIN-III C-METHYLTRANSFERASE HEMD |
| M. ulcerans Agy99 | MUL_4600 | hemD | 0.0 | 87.81% (566) | uroporphyrin-III C-methyltransferase HemD |
| M. vanbaalenii PYR-1 | Mvan_0846 | - | 0.0 | 78.96% (575) | uroporphyrinogen III synthase HEM4 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0065|M.gilvum_PYR-GCK -------------------------------------------MSLRGRK
Mvan_0846|M.vanbaalenii_PYR-1 -------------------------------------------MSLRGRK
Rv0511|M.tuberculosis_H37Rv --------------------------------------------MTRGRK
Mb0524|M.bovis_AF2122/97 --------------------------------------------MTRGRK
MAV_4638|M.avium_104 --------------------------------------------------
MMAR_0844|M.marinum_M -------------------------------------------MMTRGRK
MUL_4600|M.ulcerans_Agy99 --------------------------------------------MTRGRK
MLBr_02420|M.leprae_Br4923 --------------------------------------MVGATMTTRGRK
MAB_3991c|M.abscessus_ATCC_199 --------------------------------------------MTRGRK
MSMEG_0954|M.smegmatis_MC2_155 MGGADSLTKVGGADAAVAVVGAAEGSVGAAESPASDPQPRESSSRSRSRK
TH_0267|M.thermoresistible__bu --------------------------------------------------
Mflv_0065|M.gilvum_PYR-GCK NKPGRITFVGSGPGDPGLLTSRARAVLANAALVFTDPDVPEAVRALVGSE
Mvan_0846|M.vanbaalenii_PYR-1 GKPGRITFVGSGPGDPGLLTTRARAVLAHAALVFTDPDVPEAVLELVGSE
Rv0511|M.tuberculosis_H37Rv PRPGRIVFVGSGPGDPGLLTTRAAAVLANAALVFTDPDVPEPVVALIGTD
Mb0524|M.bovis_AF2122/97 PRPGRIVFVGSGPGDPGLLTTRAAAVLANAALVFTDPDVPEPVVALIGTD
MAV_4638|M.avium_104 --------MGSGPGDPGLLTARAAAVLANAALVFTDPDVPEPVLALIGKD
MMAR_0844|M.marinum_M PRPGRITYVGSGPGDPGLLTTRAAAVLANAALVFTDPDVPESVLALIGND
MUL_4600|M.ulcerans_Agy99 PRPGRITYVGSGPGDPGLLTTRAAAVLANAALVFTDPDVPESVLALIGND
MLBr_02420|M.leprae_Br4923 LRPGRITFVGAGPGAPGLLTAQAVAVLANAALVFIDPDVPEPVLALIGKD
MAB_3991c|M.abscessus_ATCC_199 -KPGRILFVGSGPGDPGLLTVRARAVLTGATVAFTDPDVPQAVLDLVG--
MSMEG_0954|M.smegmatis_MC2_155 VKPGRITFVGSGPGDPGLLTTRAQAVLAHAELVFTDPDVPEAVLALVGTD
TH_0267|M.thermoresistible__bu --------VGAGPGDPGLLTTKAQTILAHAALVFTDPDVPEAVKTLIGTN
:*:*** ***** :* ::*: * :.* *****:.* *:*
Mflv_0065|M.gilvum_PYR-GCK LPPPSGPLPP------VAD-AAPKTAAAGADQPASQDAAPADAAKTAKAA
Mvan_0846|M.vanbaalenii_PYR-1 LPPPSGPLPP------VAAGAAPKPADAVVKQDADAAEKPAEGSK---AA
Rv0511|M.tuberculosis_H37Rv LPPVSGPAP-----------AEPVAGNGDAAGGGSA-QEHGRAAS-----
Mb0524|M.bovis_AF2122/97 LPPVSGPAP-----------AEPVAGNGDAAGGGSA-QEHGRAAS-----
MAV_4638|M.avium_104 LPPVSGPAP-----------AEPAP-NGEGAG-----SEQGQAQP-----
MMAR_0844|M.marinum_M LPPVSGPAP-----------ATAVADNADAATGGPAEGESDQAVP-----
MUL_4600|M.ulcerans_Agy99 LPPVSGPAP-----------ATAVADNADAATGGPAEGESDQAVP-----
MLBr_02420|M.leprae_Br4923 LPPVSGPAP-----------AGPAAVTAGRAADGTLEGDHYQAAP-----
MAB_3991c|M.abscessus_ATCC_199 -SAVELPEP-----------APAKPATGGDEAPAAADNEAAAEPP-----
MSMEG_0954|M.smegmatis_MC2_155 LPPAAGPAPA------KKNDKADKATDKSDKSDKADKADDAKSDD-----
TH_0267|M.thermoresistible__bu LPPLVGPAPARPVKSGAKSDAKAAGAKSDAKATDAKSDAKAAKADDKDAV
.. * *
Mflv_0065|M.gilvum_PYR-GCK EDAVIPGGPDVRPALGDPAEVAKTLAHEARTGVDVVRLVAGDPLSNDSVI
Mvan_0846|M.vanbaalenii_PYR-1 EDAVIPGGPDVRPALGDPVEVAKTLATEARTGVDVVRLVAGDPLSIDSVI
Rv0511|M.tuberculosis_H37Rv --AVVSGGPDIRPALGDPADVAKTLTAEARSGVDVVRLVAGDPLTVDAVI
Mb0524|M.bovis_AF2122/97 --AVVSGGPDIRPALGDPADVAKTLTAEARSGVDVVRLVAGDPLTVDAVI
MAV_4638|M.avium_104 --AVISGGPDIRPALGDPAEVAKTLTAEARTGVDVVRLVAGDPLTVDAVI
MMAR_0844|M.marinum_M --AVVSSGPDIRPALGDPTEVAKTLTAEARSGADVVRLVAGDPMSVDAVI
MUL_4600|M.ulcerans_Agy99 --AVVSSGPDIRPALGDPTEVAKTLTAEARSGADVVRLVAGDPMSVDAVI
MLBr_02420|M.leprae_Br4923 --MVMSGGADIRPALGDPTEVAKTLTAEARSGVDVVRLVAGDPLTVDAVI
MAB_3991c|M.abscessus_ATCC_199 --LTI----DIRPALGDPADVAKTLVAEARSGVDVVRLVAGDPLSIDSVL
MSMEG_0954|M.smegmatis_MC2_155 -PAVIPGGPEVRPALGDPAEVAKTLASEARHGYNVVRLIAGDPLSVDAVI
TH_0267|M.thermoresistible__bu EEEDICRGPEIRPALGDPAEVAKTVAAEARSGSDVVRLIAGDPLSVDAVI
: ::*******.:****:. *** * :****:****:: *:*:
Mflv_0065|M.gilvum_PYR-GCK TEVNALAKTQLSFEIVPGLPGTSAVPTYAGLPLGSSHTVADVRG---DVD
Mvan_0846|M.vanbaalenii_PYR-1 TEINALAKTQLNFEIVPGLPGTSAVPTYAGLPLGSSHTVADVRDP--NVD
Rv0511|M.tuberculosis_H37Rv SEVNAVARTHLHIEIVPGLAASSAVPTYAGLPLGSSHTVADVRIDPENTD
Mb0524|M.bovis_AF2122/97 SEVNAVARTHLHIEIVPGLAASSAVPTYAGLPLGSSHTVADVRIDPENTD
MAV_4638|M.avium_104 TEVNAIARSHLHIEIVPGLAPTNAVPTYAGLPLGSSHTMADVRGD---VD
MMAR_0844|M.marinum_M AEVNAITRSHLQIEILPGLAATSAVPTYAGLPLGSAHTVADVRGD---VD
MUL_4600|M.ulcerans_Agy99 AEVNAITRSHLQIEILPGLAATSAVPTYAGLPLGSAHTVADVRGD---VD
MLBr_02420|M.leprae_Br4923 AEVNAVARTNLQIEIVPGLVTSSVVPTYAGLPLGSSHTVADVRGD---VD
MAB_3991c|M.abscessus_ATCC_199 AEVHAVTRTQVAFEILPGLASSTAVPAYAGLPPGSAHTVADVRGD---VD
MSMEG_0954|M.smegmatis_MC2_155 TEINALAKTHLTFEIVPGLPDTTAVPTYAGLPLGSSHTVADVRGD---VD
TH_0267|M.thermoresistible__bu NEITALARTQLTFEIVPGLPQTTAVPTYAGLPLGSTHTVADVRGD---VD
*: *:::::: :**:*** :..**:***** **:**:**** .*
Mflv_0065|M.gilvum_PYR-GCK WAALAAAPGPLILHATPSHLPEAARTLIEYGLAESTPAVVTANGTTCQQR
Mvan_0846|M.vanbaalenii_PYR-1 WAALAAAPGPLILHATASHLPEAARTLIEYGLADSTPAVVTANGTTCQQR
Rv0511|M.tuberculosis_H37Rv WDALAAAPGPLILQATASHLAESARSLIDHQLAESTPCVVTAHGTTCQQR
Mb0524|M.bovis_AF2122/97 WDALAAAPGPLILQATASHLAESARSLIDHQLAESTPCVVTAHGTTCQQR
MAV_4638|M.avium_104 WEALAAAPGPLILQATASHLADAARTLIEHELAENTPCVVTAHGTTCQQR
MMAR_0844|M.marinum_M WEALAAAPGPLILQATASHLADAARTLVEHELADTTPCVVTAHGTTCQQR
MUL_4600|M.ulcerans_Agy99 WEALAAAPGPLILQATASHLADAARTLVEHELADTTPCVVTAHGTTCQQR
MLBr_02420|M.leprae_Br4923 WEALAAAPGPLILQATASHLADAAHILIDHGLDDATPCVVTAQGTTCQQR
MAB_3991c|M.abscessus_ATCC_199 WAALAAAPGPLILHATPTHVPDAAKALIEHGLADQTLVAVTAQGTTCAQR
MSMEG_0954|M.smegmatis_MC2_155 WAALAAAPGPLILHATASHLPDAARTLIEHGLVENTPAVVTANGTTCQQR
TH_0267|M.thermoresistible__bu WGALAAAPGPLILQATASHLSDAANTLIEYGLADTTPVVVTTNGTTCQQR
* ***********:**.:*:.::*. *::: * : * .**::**** **
Mflv_0065|M.gilvum_PYR-GCK SVETTLAGLLDKAALAG------TEPAGPLTG------TLVVTIGRTVAN
Mvan_0846|M.vanbaalenii_PYR-1 SIETTLGGLLDKAVLDKP---AVAEPAGPLTG------TLVVTLGRTVAH
Rv0511|M.tuberculosis_H37Rv SVETTLQGLTDPAVLGA------TDPACSANGRDSQAGPLIVTIGKTVTS
Mb0524|M.bovis_AF2122/97 SVETTLQGLTDPAVLGA------TDPACSANGRDSQAGPLIVTIGKTVTS
MAV_4638|M.avium_104 SIETTLQGLTDSAVLGG------TDPAGPLTG------PLVVTIGKTVSA
MMAR_0844|M.marinum_M SVETTLQGLTDPAILGG------TDPAGPLTG------PLVVTIGKTVSN
MUL_4600|M.ulcerans_Agy99 SVETTLQGLTDPAILGG------TDPAGPLTG------PLVVTIGKTVSN
MLBr_02420|M.leprae_Br4923 SVETTLEGLADSAVLVG------TDTAGPLTG------PLVVTIGKTVTS
MAB_3991c|M.abscessus_ATCC_199 TVETTLASLIDGVPVDA------NDPHGPMTG------ALVLTIGRVVAG
MSMEG_0954|M.smegmatis_MC2_155 SVETTLVGLTDKTVVEKP---AGSELLGPLTG------PLVVTIGKTVAN
TH_0267|M.thermoresistible__bu SVEASLATMGDKATLAGLEPPGGAPAEGPHTG------NLVVTIGKTVTN
::*::* : * . : . .* *::*:*:.*:
Mflv_0065|M.gilvum_PYR-GCK RAKLNWWESRALYGWTVLVPRTKDQAGEMSDRLVGHGALPIEVPTIAVEP
Mvan_0846|M.vanbaalenii_PYR-1 RAKLNWWESRALYGWTVLVPRTKDQAGEMSDRLVGHGALPIEVPTIAVEP
Rv0511|M.tuberculosis_H37Rv RAKLNWWESRALYGWTVLVPRTKDQAGEMSERLTSYGALPVEVPTIAVEP
Mb0524|M.bovis_AF2122/97 RAKLNWWESRALYGWTVLVPRTKDQAGEMSERLTSYGALPVEVPTIAVEP
MAV_4638|M.avium_104 RAKLNWWESRALYGWTVLVPRTKDQAGEMSERLTSYGALPIEVPTIAVEP
MMAR_0844|M.marinum_M RAKLNWWESRALYGWTVLVPRTKDQAGEMSERLTSYGALPVEVPTIAVEP
MUL_4600|M.ulcerans_Agy99 RAKLNWWESRALYGWTVLVPRTKDQAGEMSERLTSYGALPVEVPTIAVAP
MLBr_02420|M.leprae_Br4923 RARLNWWESRALYGWTVLVPRTKDQAGEMSERLTSYGALPVEVPTIAVEP
MAB_3991c|M.abscessus_ATCC_199 RSKLNWWESRALYGWTVLVPRTKDQAGEMSDKLVGYGALPVEVPTIAVEP
MSMEG_0954|M.smegmatis_MC2_155 RAKLNWWESRALYGWTVLVPRTKDQAGEMSERLVTHGALPIEVPTIAVEP
TH_0267|M.thermoresistible__bu RAKLNWWESRALYGWTVLVPRTREQAGEMSEKLVAHGALPMEVPTISVEP
*::*******************::******::*. :****:*****:* *
Mflv_0065|M.gilvum_PYR-GCK PRSPAQMERAVKGLVDGRFQWVVFTSTNAVRAVWEKFNEFGLDARAFSGV
Mvan_0846|M.vanbaalenii_PYR-1 PRSPAQMERAVKGLVDGRFQWVVFTSTNAVRAVWEKFNEFGLDARAFSGV
Rv0511|M.tuberculosis_H37Rv PRSPAQMERAVKGLVDGRFQWIVFTSTNAVRAVWEKFGEFGLDARAFSGV
Mb0524|M.bovis_AF2122/97 PRSPAQMERAVKGLVDGRFQWIVFTSTNAVRAVWEKFGEFGLDARAFSGV
MAV_4638|M.avium_104 PRSPAQMERAVKGLVDGRFQWVVFTSTNAVRAVWEKFGEFGLDARAFSGV
MMAR_0844|M.marinum_M PRSPAQMERAVKGLVDGRFQWVVFTSTNAVRAVWEKFGEFGLDARAFSGV
MUL_4600|M.ulcerans_Agy99 PRSPAQMERAVKGLVDGRFQWVVFTSTNAVRAVWEKFGEFGLDARAFSGV
MLBr_02420|M.leprae_Br4923 PRSPAQMERAVKGLVDGRFQWVVFTSTNAVRAVWEKFGEFGLDARAFSGV
MAB_3991c|M.abscessus_ATCC_199 PRSPAQMERAVKGLVDGRYQWVVFTSTNAVRAVWEKFAEFGLDARAFSGV
MSMEG_0954|M.smegmatis_MC2_155 PRSPAQMERAVKGLVDGRFQWVVFTSTNAVRAVWEKFNEFGLDARAFSGV
TH_0267|M.thermoresistible__bu PRSPAQMERAVKGLVDGRYQWVVFTSTNSVRAVWEKFKEFGLDARAFSGV
******************:**:******:******** ************
Mflv_0065|M.gilvum_PYR-GCK KIACVGQATAEKVRAFGINPELVPTGEQSSLGLLDEFPPYDDIFDPVNRV
Mvan_0846|M.vanbaalenii_PYR-1 KIACVGQATAERVRAFGINPELVPTGEQSSLGLLDEFPPYDDIFDPVNRV
Rv0511|M.tuberculosis_H37Rv KIACVGESTADRVRAFGISPELVPSGEQSSLGLLDDFPPYDSVFDPVNRV
Mb0524|M.bovis_AF2122/97 KIACVGESTADRVRAFGISPELVPSGEQSSLGLLDDFPPYDSVFDPVNRV
MAV_4638|M.avium_104 KIACVGESTADRVRAFGISPELVPAGEQSSLGLLDDFPLYDSVFDPVNRV
MMAR_0844|M.marinum_M KIACVGESTADRVRAFGISPELVPAGEQSSLGLLDEFPPYDSIFDPVNRV
MUL_4600|M.ulcerans_Agy99 KIACVGESTADRVRAFGISPELVPAGEQSSLGLLDEFPPYDSIFDPVNRV
MLBr_02420|M.leprae_Br4923 KIACVGEATADRVRAFGISPELVPAGEQSSLGLLDEFPPYDSVFDPVNRV
MAB_3991c|M.abscessus_ATCC_199 KIACVGQATADKVRAFGINPELVPAGEQSSLGLLDEFPPYDDVFDPVNRV
MSMEG_0954|M.smegmatis_MC2_155 KIACVGQATADKVRAFGINPELVPSGEQSSLGLLDEFPPYDEVFDPVNRV
TH_0267|M.thermoresistible__bu KIACVGQATADRVRAFGINPELVPSGEQSSLGLLEEFPPYDDVFDPVNRV
******::**::******.*****:*********::** **.:*******
Mflv_0065|M.gilvum_PYR-GCK LLPRADIATETLAEGLRELGWEIEDVTAYRTVRAAPPPAHTREMIKTGGF
Mvan_0846|M.vanbaalenii_PYR-1 LLPRADIATETLAEGLRERGWEIEDVTAYRTVRAAPPPAQTREMIKTGGF
Rv0511|M.tuberculosis_H37Rv LLPRADIATETLAEGLRERGWEIEDVTAYRTVRAAPPPATTREMIKTGGF
Mb0524|M.bovis_AF2122/97 LLPRADIATETLAEGLRERGWEIEDVTAYRTVRAAPPPATTREMIKTGGF
MAV_4638|M.avium_104 LLPRADIATETLAEGLRERGWEIEDVTAYRTVRAAPPPAETREMIKTGGF
MMAR_0844|M.marinum_M LLPRADIATETLAEGLRERGWEIEDVTAYRTVRAAPPPAATREMIKTGGF
MUL_4600|M.ulcerans_Agy99 LLPRADIATETLAEGLRERGWEIEDVTAYRTVRAAPPPAATREMIKTGGF
MLBr_02420|M.leprae_Br4923 LLPRADIATETLAEGLRGCGWEIEDVTAYRTVRAAPPPAATREMIKTGGF
MAB_3991c|M.abscessus_ATCC_199 LLPRADIATETLAEGLRERGWEIDDVTAYRTVRAAPPAAQTREMIKTGGF
MSMEG_0954|M.smegmatis_MC2_155 LLPRADIATETLAEGLRERGWEIEDVTAYRTVRAAPPPAHTREMIKTGGF
TH_0267|M.thermoresistible__bu LLPRADIAXXX---------------------------------------
********
Mflv_0065|M.gilvum_PYR-GCK DAVCFTSSSTVRNLVGIAGKPHARTIVACIGPKTAETAAEFGLRVDVQPE
Mvan_0846|M.vanbaalenii_PYR-1 DAVCFTSSSTVRNLVGIAGKPHARTIVACIGPKTAETAAEFGLRVDVQPE
Rv0511|M.tuberculosis_H37Rv DAVCFTSSSTVRNLVGIAGKPHARTIIACIGPKTAETAAEFGLRVDVQPD
Mb0524|M.bovis_AF2122/97 DAVCFTSSSTVRNLVGIAGKPHARTIIACIGPKTAETAAEFGLRVDVQPD
MAV_4638|M.avium_104 DAVCFTSSSTVRNLVGIAGKPHARTIIACIGPKTAETAAEFGLRVDVQPE
MMAR_0844|M.marinum_M DAVCFTSSSTVRNLVGIAGKPHARTIIACIGPKTAETAAEFGLRVDVQPE
MUL_4600|M.ulcerans_Agy99 DAVCFTSSSTVRNLVGIAGKPHARTIIACIGPKTAETAAEFGLRVDVQPE
MLBr_02420|M.leprae_Br4923 DAVCFTSSSTVRNLVGIAGKPHARTVIACIGPKTAETASEFGLRVDVQPE
MAB_3991c|M.abscessus_ATCC_199 DAVCFTSSSTVRNLVGIAGKPHARTIVACIGPKTAETAVEFGLRVDVQPE
MSMEG_0954|M.smegmatis_MC2_155 DAVCFTSSSTVRNLVGIAGKPHARTIVACIGPKTAETAIEFGLRVDVQPE
TH_0267|M.thermoresistible__bu --------------------------------------------------
Mflv_0065|M.gilvum_PYR-GCK VAAVGPLVEALAEHAARLRAEGALPPPRKKSRRR
Mvan_0846|M.vanbaalenii_PYR-1 VAAVGPLVEALAEHAARLRAEGALPPPRKKSRRR
Rv0511|M.tuberculosis_H37Rv TAAIGPLVDALAEHAARLRAEGALPPPRKKSRRR
Mb0524|M.bovis_AF2122/97 TAAIGPLVDALAEHAARLRAEGALPPPRKKSRRR
MAV_4638|M.avium_104 TAAVGPLVDALAEHAARLRAEGALPPPRKKSRRR
MMAR_0844|M.marinum_M TAAVGPLVDALAEHAARLRAEGALPPPRKKSRRR
MUL_4600|M.ulcerans_Agy99 TAAVGPLVDALAEHAARLRAEGALPPPRKKSRRR
MLBr_02420|M.leprae_Br4923 TAAVGPLVDALAEHVARLRAEGALPPPRKKSRRR
MAB_3991c|M.abscessus_ATCC_199 TAAVGPLVEALAEHAARLRTEGALPPPRKKSRRR
MSMEG_0954|M.smegmatis_MC2_155 TAAVGPLVEALAEHAARLRAEGALPPPRKKSRRR
TH_0267|M.thermoresistible__bu ----------------------------------