For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TENPYLVGLRLAGKKVVVVGGGTVAQRRLPLLIASGADVHVIAPSVTPAVEAMDQITLSVRDYRDGDLDG AWYAIAATDDARVNVAVVAEAERRRIFCVRADIAVEGTAVTPASFSYAGLSVGVLAGGEHRRSAAIRSAI REALQQGVITAQSSDVLSGGVALVGGGPGDPELITVRGRRLLAQADVVVADRLAPPELLAELPPHVEVID AAKIPYGRAMAQDAINAVLIERARSGNFVVRLKGGDPFVFARGYEEVLACAHAGIPVTVVPGVTSAIAVP AMAGVPVTHRAMTHEFVVVSGHLAPGHPESLVNWDALAALTGTIVLLMAVERIELFVDVLLKGGRTADTP VLVVQHGTTAAQQTLRATLADTPEKVRAAGIRPPAIIVIGAVVGLSGVRGLNNS*
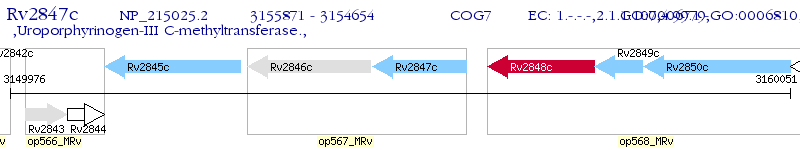
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2847c | cysG | - | 100% (405) | POSSIBLE MULTIFUNCTIONAL ENZYME SIROHEME SYNTHASE CYSG: UROPORPHYRIN-III C-METHYLTRANSFERASE (UROGEN III METHYLASE) (SUMT) (UROPORPHYRINOGEN III METHYLASE) (UROM) + PRECORRIN-2 OXIDASE + FERROCHELATASE |
| M. tuberculosis H37Rv | Rv2071c | cobM | 2e-24 | 34.16% (243) | precorrin-4 C11-methyltransferase CobM |
| M. tuberculosis H37Rv | Rv0511 | hemD | 5e-19 | 29.50% (261) | uroporphyrin-III C-methyltransferase HemD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2872c | cysG | 0.0 | 100.00% (405) | multifunctional uroporphyrinogen III methylase/precorrin-2 |
| M. gilvum PYR-GCK | Mflv_4047 | - | 0.0 | 82.07% (396) | uroporphyrin-III C-methyltransferase |
| M. leprae Br4923 | MLBr_02420 | hemD | 3e-19 | 28.87% (284) | putative uroporphyrin-III C-methyltransferase |
| M. abscessus ATCC 19977 | MAB_3143c | - | 1e-172 | 78.88% (393) | multifunctional siroheme synthase CysG/uroporphyrin-III |
| M. marinum M | MMAR_1886 | cysG | 0.0 | 85.35% (396) | multifunctional enzyme siroheme synthase CysG |
| M. avium 104 | MAV_3703 | cobA | 0.0 | 88.64% (396) | uroporphyrin-III C-methyltransferase |
| M. smegmatis MC2 155 | MSMEG_2618 | cobA | 0.0 | 80.85% (402) | uroporphyrin-III C-methyltransferase |
| M. thermoresistible (build 8) | TH_1981 | cysG2 | 0.0 | 80.68% (409) | POSSIBLE MULTIFUNCTIONAL ENZYME SIROHEME SYNTHASE CYSG: |
| M. ulcerans Agy99 | MUL_2114 | cysG | 0.0 | 85.10% (396) | multifunctional enzyme siroheme synthase CysG |
| M. vanbaalenii PYR-1 | Mvan_2305 | - | 0.0 | 81.36% (397) | uroporphyrin-III C-methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2847c|M.tuberculosis_H37Rv --MTENPYLVGLRLAGKKVVVVGGGT----VAQRRLPLLIASGADVHVIA
Mb2872c|M.bovis_AF2122/97 --MTENPYLVGLRLAGKKVVVVGGGT----VAQRRLPLLIASGADVHVIA
MMAR_1886|M.marinum_M --MTESPYLVGLRLTGRKVVVVGGGT----VAQRRLPLLIASGADVQVIS
MUL_2114|M.ulcerans_Agy99 --MTESPYLVGLRLTGRKVVVVGGGT----VAQRRLPLLIASGADVQVIS
MAV_3703|M.avium_104 --MTESAYLVGLRLTGKKVVVVGGGT----VAQRRLPLLIASGADVHVIS
MSMEG_2618|M.smegmatis_MC2_155 --MTEDAYLVGLRLSGKKVVVVGGGT----VAQRRLPLLLANGADVHVIT
TH_1981|M.thermoresistible__bu --VSENAYLAGLLLSGKKVIVVGGGG----VAQRRLPLLIASGADVHVIA
Mflv_4047|M.gilvum_PYR-GCK MSTYDNAYLVGLRLSGKKVVVVGGGT----VAQRRLPLLVSNGADVHVVT
Mvan_2305|M.vanbaalenii_PYR-1 -MTSENAYLVGLRLAGKKVVVVGGGT----VAQRRLPLLVASGADVHVIT
MAB_3143c|M.abscessus_ATCC_199 --MTHDAYLVGLRLAGRKVVVVGAGS----VAQRRLGLLIASGADVHVIA
MLBr_02420|M.leprae_Br4923 -MVGATMTTRGRKLRPGRITFVGAGPGAPGLLTAQAVAVLANAALVFIDP
* * :: .**.* : : :::..* * : .
Rv2847c|M.tuberculosis_H37Rv PSVTPAVEAMD-------------QITLSVRDYRDGDLDGAWYAIA----
Mb2872c|M.bovis_AF2122/97 PSVTPAVEAMD-------------QITLSVRDYRDGDLDGAWYAIA----
MMAR_1886|M.marinum_M RSATRSVEVMD-------------KITLTLREYRDGDLEGAWYAIA----
MUL_2114|M.ulcerans_Agy99 RSATRSVEVMD-------------KITLTLREYRDGDLEGAWYAIA----
MAV_3703|M.avium_104 RSATRSVEAMT-------------GITLQLREYRDGDLDGAWYAIA----
MSMEG_2618|M.smegmatis_MC2_155 RAATPVVEALQKDDP---------GITLELRDFRPGDLEGAWYVIA----
TH_1981|M.thermoresistible__bu RSATPAVEALSRQQP---------GITLELREFRDGDLEGAWYVIA----
Mflv_4047|M.gilvum_PYR-GCK KAATPAVEAID-------------GITLTLREFRDEDLDGAWYVIA----
Mvan_2305|M.vanbaalenii_PYR-1 RAATPAVEAID-------------GITLTMRDFRAGDLEGAWYVIA----
MAB_3143c|M.abscessus_ATCC_199 PHATPAVEGMA-------------SINLQLRPYQDGDLDGAWYAIA----
MLBr_02420|M.leprae_Br4923 DVPEPVLALIGKDLPPVSGPAPAGPAAVTAGRAADGTLEGDHYQAAPMVM
: : : *:* * *
Rv2847c|M.tuberculosis_H37Rv --------ATDDARVNVAVVAEAERRRIFCVR----ADIAVEGTAVTPAS
Mb2872c|M.bovis_AF2122/97 --------ATDDARVNVAVVAEAERRRIFCVR----ADIAVEGTAVTPAS
MMAR_1886|M.marinum_M --------ATDDPAVNAAIVAEADRRQIFCVR----ADIAVEGTAVTPAT
MUL_2114|M.ulcerans_Agy99 --------ATDDPAVNAAIVAEADRRQIFCVR----ADIAVEGTAVTPAT
MAV_3703|M.avium_104 --------ATDDPRVNEAVVAEAERRRIFCVR----ADVAVAGTAVTPAS
MSMEG_2618|M.smegmatis_MC2_155 --------ATDDPVVNAAIAAEADERRIFCVR----ADIAREGSAVTPAT
TH_1981|M.thermoresistible__bu --------ATDDPDVNASVVAEADRRRIFCVR----ADVAREGSAVTPAT
Mflv_4047|M.gilvum_PYR-GCK --------ATDDPAVNAAIVAGAERRRIFCVR----ADIAVEGTAVTPAT
Mvan_2305|M.vanbaalenii_PYR-1 --------ATDDPEVNAAIVAEAESRRVFCVR----ADVAVEGTAVTPAT
MAB_3143c|M.abscessus_ATCC_199 --------CTDDPAVNAAVVDEAERRHIFCVR----ADSARDGTAVTPAS
MLBr_02420|M.leprae_Br4923 SGGADIRPALGDPTEVAKTLTAEARSGVDVVRLVAGDPLTVDAVIAEVNA
. .*. : ** : . . :
Rv2847c|M.tuberculosis_H37Rv FSYAGLSVGVLAG---------------GEHRRSAAIRSAIREALQQGVI
Mb2872c|M.bovis_AF2122/97 FSYAGLSVGVLAG---------------GEHRRSAAIRSAIREALQQGVI
MMAR_1886|M.marinum_M FNHAGLSVGVLAS---------------GDHRRSAAIRSAIREALQNGLV
MUL_2114|M.ulcerans_Agy99 FNHAGLSVGVLAS---------------GDHRRSAAIRSAIREALQNGLV
MAV_3703|M.avium_104 FSYAGLSVGVLAG---------------GEHRRSAAIRSAIREALQTGVI
MSMEG_2618|M.smegmatis_MC2_155 FEHDGLSVGVLAG---------------GEHRRSAAIRSAIHEALQQGLV
TH_1981|M.thermoresistible__bu FEYEGLSVGVLAG---------------GEHRRSAAIRSAIREALQQGVI
Mflv_4047|M.gilvum_PYR-GCK FEYEGLEVGVLAG---------------GEHRRSAAIRTAIREALQQGVV
Mvan_2305|M.vanbaalenii_PYR-1 FEYEGLSVGVLAG---------------GEHRRSAGIRTAIREALQRGLI
MAB_3143c|M.abscessus_ATCC_199 FNHDGIAVGVLTG---------------GQHKRSAALRSAIHEALQRGLI
MLBr_02420|M.leprae_Br4923 VARTNLQIEIVPGLVTSSVVPTYAGLPLGSSHTVADVRGDVDWEALAAAP
. .: : ::.. *. : * :* : .
Rv2847c|M.tuberculosis_H37Rv TAQ-----SSDVLSGGVALVGGGPGDPE--LITVRGRRLLAQADVVVADR
Mb2872c|M.bovis_AF2122/97 TAQ-----SSDVLSGGVALVGGGPGDPE--LITVRGRRLLAQADVVVADR
MMAR_1886|M.marinum_M TSTSYQ--SSDVIRGGVALIGGGPGDPE--LITVRGRRLLAQADVVVADR
MUL_2114|M.ulcerans_Agy99 TSTSYQ--SSDVIRGGVALIGGGPGDPE--LITVRGRRLLAQADVVVADR
MAV_3703|M.avium_104 TPEGPV--GSDAVKGGVALIGGGPGDPE--LITVRGRRLLAQADVVVADR
MSMEG_2618|M.smegmatis_MC2_155 TED-----MPGELTSGVALVGGGPGDPE--LITVRGRRLLARADVVVADR
TH_1981|M.thermoresistible__bu TAEAGADPMSGAIPGGVALVGGGPGDPE--LITVRGRRLLAQADVVVADR
Mflv_4047|M.gilvum_PYR-GCK TDESGD--APGVVKGGVALVGGGPGDPE--LITVRGRRLLAHADVVVADR
Mvan_2305|M.vanbaalenii_PYR-1 GVDSSA--APGVAKGTVALVGGGPGDPE--LVTVRGRRLLAHADVVVADR
MAB_3143c|M.abscessus_ATCC_199 TET------ADVKPEGVALVGGGPGDPD--LITVRGRRLLAQADVVVADR
MLBr_02420|M.leprae_Br4923 GPLILQATASHLADAAHILIDHGLDDATPCVVTAQGTTCQQRSVETTLEG
. *:. * .*. ::*.:* :: .. :
Rv2847c|M.tuberculosis_H37Rv LAPPELLAELPPHVEVIDAAKIPYGRAMAQDAIN-----------AVLIE
Mb2872c|M.bovis_AF2122/97 LAPPELLAELPPHVEVIDAAKIPYGRAMAQDAIN-----------AVLIE
MMAR_1886|M.marinum_M LAPPELLAELPPHVEVIDAAKIPYGRAMAQDAIN-----------DVMIE
MUL_2114|M.ulcerans_Agy99 LAPPELLAELPPHVEVIDAAKIPYGRAMAQDAIN-----------DVMIE
MAV_3703|M.avium_104 LAPPELLAELPPHVEVIDAAKIPYGRAMAQDAIN-----------DVMIE
MSMEG_2618|M.smegmatis_MC2_155 LAPQELLAELGPHVEVIDAAKIPYGRAMAQEAIN-----------AVLIE
TH_1981|M.thermoresistible__bu LAPQELLAELPPHVEVIDAAKIPYGRAMAQEAIN-----------ATLID
Mflv_4047|M.gilvum_PYR-GCK LAPQELLAELSPEVEVIDASKIPYGRAMAQDAIN-----------AVLVD
Mvan_2305|M.vanbaalenii_PYR-1 LAPQDLLSELSPEVEVIDAAKIPYGRAMAQDAIN-----------EVLIA
MAB_3143c|M.abscessus_ATCC_199 LAPAELLADLGPHVEVIDAAKIPYGRAMAQQEIN-----------RVLIE
MLBr_02420|M.leprae_Br4923 LADSAVLVGTDTAGPLTGPLVVTIGKTVTSRARLNWWESRALYGWTVLVP
** :* . : .. :. *::::. .::
Rv2847c|M.tuberculosis_H37Rv RAR--SGNFVVRLKGGDPFVFARGYEEVLACAHAGIPVTVVPGVTS----
Mb2872c|M.bovis_AF2122/97 RAR--SGNFVVRLKGGDPFVFARGYEEVLACAHAGIPVTVVPGVTS----
MMAR_1886|M.marinum_M RAK--AGRFVVRLKGGDPFVFARGYEEVLACTEAGIPVIVVPGVTS----
MUL_2114|M.ulcerans_Agy99 RAK--AGRFVVRLKGGDPFVFARGYEEVLACTEAGIPVIVVPGVTS----
MAV_3703|M.avium_104 RAR--AGSFVVRLKGGDPFVFARGYEEVLACADAGIPVTVVPGVTS----
MSMEG_2618|M.smegmatis_MC2_155 RAR--AGKFVVRLKGGDPFVFARGYEEVLACSEAGIPVTVVPGVTS----
TH_1981|M.thermoresistible__bu RAR--AGKFVVRLKGGDPFVFARGYEEVIACAEAGIPVTVVPGITS----
Mflv_4047|M.gilvum_PYR-GCK RAK--AGKFVVRLKGGDPFVFARGYEEVIACAEAGIPVTVVPGVTS----
Mvan_2305|M.vanbaalenii_PYR-1 RAR--EGKFVVRLKGGDPFVFARGYEEVIACAEAGIPVTVVPGVTS----
MAB_3143c|M.abscessus_ATCC_199 RAQ--EGKFVVRLKGGDPFVFARGYEEVLACAEAGVQVTVVPGVTS----
MLBr_02420|M.leprae_Br4923 RTKDQAGEMSERLTSYGALPVEVPTIAVEPPRSPAQMERAVKGLVDGRFQ
*:: * : **.. ..: . * . .. .* *:..
Rv2847c|M.tuberculosis_H37Rv --------------------AIAVPAMAGVPVTHR-----------AMTH
Mb2872c|M.bovis_AF2122/97 --------------------AIAVPAMAGVPVTHR-----------AMTH
MMAR_1886|M.marinum_M --------------------AIAVPALAGVPVTHR-----------ATNH
MUL_2114|M.ulcerans_Agy99 --------------------AIAVPALAGVPVTHR-----------ATNH
MAV_3703|M.avium_104 --------------------AIGVPALAGVPVTHR-----------AVNH
MSMEG_2618|M.smegmatis_MC2_155 --------------------AIAVPALAGVPVTHR-----------GMTH
TH_1981|M.thermoresistible__bu --------------------AIGVPALAGVPVTHR-----------AVNH
Mflv_4047|M.gilvum_PYR-GCK --------------------AIAVPALAGVPVTHR-----------GLTH
Mvan_2305|M.vanbaalenii_PYR-1 --------------------AIAVPALAGVPVTHR-----------GLTH
MAB_3143c|M.abscessus_ATCC_199 --------------------AISVPAAAGVPVTHR-----------AVNH
MLBr_02420|M.leprae_Br4923 WVVFTSTNAVRAVWEKFGEFGLDARAFSGVKIACVGEATADRVRAFGISP
.: . * :** :: . .
Rv2847c|M.tuberculosis_H37Rv EFVVVSGHLAPGHPESLVNWDALAALTGTIVLLMAVERIELFVDVLLKGG
Mb2872c|M.bovis_AF2122/97 EFVVVSGHLAPGHPESLVNWDALAALTGTIVLLMAVERIELFVDVLLKGG
MMAR_1886|M.marinum_M EFVVVSGHIAPDHPESLVNWNALAAMSGTIVLLMAVERIELFVEALLKGG
MUL_2114|M.ulcerans_Agy99 EFVVVSGHIAPDHPESLVNWNALAAMSGTIVLLMAVERIELFVEALLKGG
MAV_3703|M.avium_104 EFVVVSGHLPPGHPESLVNWDALAALTGTIVLLMAVERIELFAEALLKGG
MSMEG_2618|M.smegmatis_MC2_155 EFVVVSGHVAPGHPESLVNWDALAAMRGTIVLLMAVERIEQFAKVLLEGG
TH_1981|M.thermoresistible__bu EFVVVSGHVAPGEPESLTNWDALAKLSGTIVLLMAVERIEVFAKALLEGG
Mflv_4047|M.gilvum_PYR-GCK EFVVVSGHLAPDHPESLVNWDALAAMSGTIVLLMAVERIELFVKVLLKGG
Mvan_2305|M.vanbaalenii_PYR-1 EFVVVSGHVAPDHPESLVNWDALAAMNGTIVLLMAVERIELFVKILLRGG
MAB_3143c|M.abscessus_ATCC_199 EFVVVSGHVAPGHPESLVNWDALAALNGTIVLLMAVERIEQFARVLIEGG
MLBr_02420|M.leprae_Br4923 ELVPAGEQSSLGLLDEFPPYDSVFDPVNRVLLPRADIATETLAEGLRGCG
*:* .. : . . :.: :::: . ::* * * :. * *
Rv2847c|M.tuberculosis_H37Rv ------------RTADTPVLVVQHGTTAAQQTLRATLADTPEKVRAAGIR
Mb2872c|M.bovis_AF2122/97 ------------RTADTPVLVVQHGTTAAQQTLRATLADTPEKVRAAGIR
MMAR_1886|M.marinum_M ------------RPADTPVLVVQHGTTPAQHTLRATLADTPEKVRAEGIR
MUL_2114|M.ulcerans_Agy99 ------------RPAYTPVLVVQHGTTPAQHTLRATLADTPEKVRAEGIR
MAV_3703|M.avium_104 ------------RPADTPVLVVQHGTTAAQHTLRATLADTPEKIRAEGIR
MSMEG_2618|M.smegmatis_MC2_155 ------------RPADTPVLVVQHGTTMAQRTLHATLADAPERIRSEGIR
TH_1981|M.thermoresistible__bu ------------RPADTPVLVVQHGTTAAQRTLRATLRDAPERMRAEGIR
Mflv_4047|M.gilvum_PYR-GCK ------------RPADTPVLVVQHGTTSSQRTLRAGLSEVPERIRSDGIR
Mvan_2305|M.vanbaalenii_PYR-1 ------------RPADTPVLVVQHGTTSSQRTLRATLSEAPERIRSDGIR
MAB_3143c|M.abscessus_ATCC_199 ------------RPAHTPVLVVQHGTTDEQRVLRADLATAPERIRSQGIR
MLBr_02420|M.leprae_Br4923 WEIEDVTAYRTVRAAPPPAATREMIKTGGFDAVCFTSSSTVRNLVGIAGK
*.* .*. . : .* .: . ..: . . :
Rv2847c|M.tuberculosis_H37Rv PPAIIVIGAVVGLSGVRGLNNS----------------------------
Mb2872c|M.bovis_AF2122/97 PPAIIVIGAVVGLSGVRGLNNS----------------------------
MMAR_1886|M.marinum_M PPAIIVIGAVAAFG-V----------------------------------
MUL_2114|M.ulcerans_Agy99 PPAIIVIGAVATFG-V----------------------------------
MAV_3703|M.avium_104 PPAIIVIGAVAAFG-V----------------------------------
MSMEG_2618|M.smegmatis_MC2_155 PPAIIVIGPVAGFAG-----------------------------------
TH_1981|M.thermoresistible__bu PPAIIVIGAVAGFDGLKNS-------------------------------
Mflv_4047|M.gilvum_PYR-GCK PPAIVVIGAVAGFGT-----------------------------------
Mvan_2305|M.vanbaalenii_PYR-1 PPAIIVIGAVAGFGA-----------------------------------
MAB_3143c|M.abscessus_ATCC_199 PPAIIVIGPVAAYGAF----------------------------------
MLBr_02420|M.leprae_Br4923 PHARTVIACIGPKTAETASEFGLRVDVQPETAAVGPLVDALAEHVARLRA
* * **. :
Rv2847c|M.tuberculosis_H37Rv --------------
Mb2872c|M.bovis_AF2122/97 --------------
MMAR_1886|M.marinum_M --------------
MUL_2114|M.ulcerans_Agy99 --------------
MAV_3703|M.avium_104 --------------
MSMEG_2618|M.smegmatis_MC2_155 --------------
TH_1981|M.thermoresistible__bu --------------
Mflv_4047|M.gilvum_PYR-GCK --------------
Mvan_2305|M.vanbaalenii_PYR-1 --------------
MAB_3143c|M.abscessus_ATCC_199 --------------
MLBr_02420|M.leprae_Br4923 EGALPPPRKKSRRR