For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NAVTVLLAYAVALSWFAPALLTASASAGAHPRLAVAGWIAAVGSALSAWAGAVVILVVGAVHAVVTRSAL TFCVETLGLTSAVHLPSSTATALVVVLLIATSAVALYTVRRVVMALIRGRRRNVEHAEAVSIVGRPTEHP GVVEISARQPTAYCVSGGGRSVIVMTTAVRDLLDEDGMAAVLAHERAHLRGRHHHVIAVLNALAAALPRL PLMRRAAREVAALLEMCADDAASREHGRTHLLNSLALLSTGDRLPEGALAAAGTAVLDRLLRLAQPPATS WWRPHVAVFAVMAACGVPIAALSLCVY*
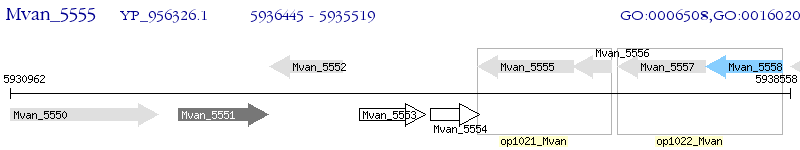
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5555 | - | - | 100% (308) | peptidase M56, BlaR1 |
| M. vanbaalenii PYR-1 | Mvan_3654 | - | 6e-52 | 40.00% (310) | hypothetical protein Mvan_3654 |
| M. vanbaalenii PYR-1 | Mvan_3707 | - | 6e-47 | 41.48% (311) | peptidase M48, Ste24p |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1876c | - | 3e-25 | 32.70% (315) | hypothetical protein Mb1876c |
| M. gilvum PYR-GCK | Mflv_0897 | - | 8e-54 | 42.57% (296) | hypothetical protein Mflv_0897 |
| M. tuberculosis H37Rv | Rv1845c | - | 3e-25 | 32.70% (315) | hypothetical protein Rv1845c |
| M. leprae Br4923 | MLBr_02064 | - | 3e-24 | 31.80% (283) | integral membrane protein |
| M. abscessus ATCC 19977 | MAB_4859 | - | 2e-45 | 38.59% (311) | hypothetical protein MAB_4859 |
| M. marinum M | MMAR_5059 | - | 9e-62 | 43.14% (306) | peptidase M48 like- protein |
| M. avium 104 | MAV_2870 | - | 1e-25 | 32.91% (313) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_3631 | - | 1e-25 | 31.15% (305) | integral membrane protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4135 | - | 3e-61 | 44.63% (307) | peptidase M48 like- protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1876c|M.bovis_AF2122/97 -MSALAFTILAVLLAGPTPALLARATWPLRAPRAAMVLWQAIALAAVLSS
Rv1845c|M.tuberculosis_H37Rv -MSALAFTILAVLLAGPTPALLARATWPLRAPRAAMVLWQAIALAAVLSS
MLBr_02064|M.leprae_Br4923 -MSALAFTILAVLLVGPTPTLVARSTWPLRAPRAAMVLWQTIALAAALST
MAV_2870|M.avium_104 -MSALAFTILAVLLTGPVPAMLARARWPLRAPRAAMVLWQAVALAAVLSA
MSMEG_3631|M.smegmatis_MC2_155 -MSALAFTLVALALVGPVPAMLARASWPLRAPRAAIVLWQSIALAAVLSA
MMAR_5059|M.marinum_M MNAALLLFAYASALIWVGPVLLRRVTGTGVHPRLAVASWLSAVFAAIAAW
MUL_4135|M.ulcerans_Agy99 MNAALLLFAYASALIWVGPVLLRRVTGTGVHPRLAVASWLSAVFAAIAAW
Mvan_5555|M.vanbaalenii_PYR-1 MNAVTVLLAYAVALSWFAPALLTASASAGAHPRLAVAGWIAAVGSALSAW
Mflv_0897|M.gilvum_PYR-GCK MTTALWLVLYGAVLAWFAPPVLRRLTGGGVSPSMGVAAWLATVAAVLVAW
MAB_4859|M.abscessus_ATCC_1997 MSIAVCLLLYSVAVLIFGPRLLRRLTRTGYAPRLAITAWLAAIVSVLGTW
. : . : * :: * .:. * : :. :
Mb1876c|M.bovis_AF2122/97 FSAGIAIASRLLMPGPDGRPTTSFVGAAGRLGWPLWAAYITVFALTVLVG
Rv1845c|M.tuberculosis_H37Rv FSAGIAIASRLLMPGPDGRPTTSFVGAAGRLGWPLWAAYITVFALTVLVG
MLBr_02064|M.leprae_Br4923 FSAGIAIASRVLMPGPDGRLTASVIGAAGRLGWPLWAAYVAAFALTVLVG
MAV_2870|M.avium_104 FSAGIAIATRVLVPGPDGRPTTSILGAEGRLGWPLWTAYIGVFALTVLVG
MSMEG_3631|M.smegmatis_MC2_155 FSAGIAIASRLFVPGADGRPTATITSEIDVLGWPLWLLYVVVFSLTLFIG
MMAR_5059|M.marinum_M VGALVFLGAAVLDSIWR-HRALALCLKELG--LAGELGLPRGIASALAVG
MUL_4135|M.ulcerans_Agy99 VGALVSLGAAVLDSIWR-HRALALCVKELG--LAGELGLPRGIASALAVG
Mvan_5555|M.vanbaalenii_PYR-1 AGAVVILVVGAVHAVVT-RSALTFCVETLG--LTSAVHLPSSTATALVVV
Mflv_0897|M.gilvum_PYR-GCK L-AAVVLVVDATSSSVLNGSVVTLCLELFG--FSDHTPLAGRLG---SIA
MAB_4859|M.abscessus_ATCC_1997 ISAAALIVIDVVRHWNSPAVVLAACAARLHAMLIGQAGAAAQVG---LLA
* : . : . :
Mb1876c|M.bovis_AF2122/97 ARLAVAVVRVATATRRRRAHHRMVVDLVGVGHNGALAQPCARARDLRVLD
Rv1845c|M.tuberculosis_H37Rv ARLAVAVVRVATATRRRRAHHRMVVDLVGVGHNGALAQPCARARDLRVLD
MLBr_02064|M.leprae_Br4923 ARLIVAIVRVAIATRRRRAHHRMVVDLVGVGHNAALAQPCARARDLRVLE
MAV_2870|M.avium_104 ARLMVAVVRVAIANRRRRAHHRMVVDLVGMGHGAALSQPCSRTRDLRVLD
MSMEG_3631|M.smegmatis_MC2_155 ARLCVAIVQVGVATRRRRAHHRMMVDLLSKSRDAVPAHLRTPASGLRILD
MMAR_5059|M.marinum_M VLSTGAVATVIACWRMGRQLRRQRSASRLHAATARTIGGASDWPGVVVVP
MUL_4135|M.ulcerans_Agy99 VLSTGAVATVIVCWRMGRQLRGQRSASRLHAATARTIGGVSDWPGVVVVP
Mvan_5555|M.vanbaalenii_PYR-1 LLIATSAVALYTVRRVVMALIRGRRRNVEHAEAVSIVGRPTEHPGVVEIS
Mflv_0897|M.gilvum_PYR-GCK LIGGGLLTSAVVALRLGRCLARLRTRSHEHAHAARIVGRPTDHPHVVVVE
MAB_4859|M.abscessus_ATCC_1997 LSAAGSIAAPALGVRLARTLIRLRDTAHEHAYAVHMVGRRTAEGDVMVLE
. * . : : :
Mb1876c|M.bovis_AF2122/97 VAQPLAYCLPGVR-SRVVVSEGTLTALADAEVAAILTHERAHLRARHDLV
Rv1845c|M.tuberculosis_H37Rv VAQPLAYCLPGVR-SRVVVSEGTLTALADAEVAAILTHERAHLRARHDLV
MLBr_02064|M.leprae_Br4923 VAQPLAYCLPGVR-SRVVVSEGALTKLNDTEVTAILTHERAHLRARHDLV
MAV_2870|M.avium_104 VPQPLAYCLPGVR-SRVVVSEGTLSTLADAEVSAILTHERAHLRARHDLV
MSMEG_3631|M.smegmatis_MC2_155 VAQPLAYCLPGVR-SRVVVSEGTLDTLSPDEVDAILTHERAHLRARHDLV
MMAR_5059|M.marinum_M APQPAAYCVTGRPNSVIVVTSGALGRLDEPQLAAVLAHEYAHLASRHHDL
MUL_4135|M.ulcerans_Agy99 APQPAAYCVTGRPNSVIVVTSGALGRLDEPQLAAVLAHEYAHLASRHHDL
Mvan_5555|M.vanbaalenii_PYR-1 ARQPTAYCVSGGGRSVIVMTTAVRDLLDEDGMAAVLAHERAHLRGRHHHV
Mflv_0897|M.gilvum_PYR-GCK ADLPAAYCVIGRP-HAIVVTSAAVRSLDRAQLKAVLAHEHAHLSGHHHHI
MAB_4859|M.abscessus_ATCC_1997 APEAAAYCVAGRP-SAIVLTTGALAVLDDAQVAAILAHERAHLAGHHPQL
. . ***: * :*:: .. * : *:*:** *** .:* :
Mb1876c|M.bovis_AF2122/97 LEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRTPLARALV
Rv1845c|M.tuberculosis_H37Rv LEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRTPLARALV
MLBr_02064|M.leprae_Br4923 LEAFTAVHAAFPRLVRSSAALSAVRLLVELLADDAAVRAAGRTPLARALV
MAV_2870|M.avium_104 LEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRAPLARALV
MSMEG_3631|M.smegmatis_MC2_155 LEMFTAVHAAFPRFVRSASALDAVRLLIELLADDAAVRIAGPTPLARALV
MMAR_5059|M.marinum_M LMALRALASSLPRLPLFAAAAKVVADLLEMCADDVAVRSHGKMALLRGLL
MUL_4135|M.ulcerans_Agy99 LMALRALASSLPRLPLFAAAAKVVADLLEMCADDVAVRSHGKMALLRGLL
Mvan_5555|M.vanbaalenii_PYR-1 IAVLNALAAALPRLPLMRRAAREVAALLEMCADDAASREHGRTHLLNSLA
Mflv_0897|M.gilvum_PYR-GCK LMVLRALAATLPHLPLFAGAHQAVAELLEMCADDTAARRVGTRPLLTGLL
MAB_4859|M.abscessus_ATCC_1997 VSVLRALADVFPRVRLMTDGSAQISRLLEMCADDSAARHHGRGVLLSGLM
: : *: :*:. . : *:*: *** * * * * .*
Mb1876c|M.bovis_AF2122/97 ACASG-RAPSGALAVGGPSTVLRVRRLSGRGNSAVLSAAAYLAAAAVLVV
Rv1845c|M.tuberculosis_H37Rv ACASG-RAPSGALAVGGPSTVLRVRRLSGRGNSAVLSAAAYLAAAAVLVV
MLBr_02064|M.leprae_Br4923 ACASG-QAPSGALAAGGNTTVLRVRRLSGRSNSAVVSAAAYLAAAIVLLV
MAV_2870|M.avium_104 ACASG-RAPSGALAAGGPSTVVRVRRLGGRGNSPLLSAAAYLAAAAVFVV
MSMEG_3631|M.smegmatis_MC2_155 TCAAG-RTPSGALAAGGPTTVIRVRRLGGKPNSLTLSVIAYLAAVAILVV
MMAR_5059|M.marinum_M ALTEGPAMAPAALGAANTATVARAERLAISVSRRSWWQERLMLWATIGLA
MUL_4135|M.ulcerans_Agy99 ALTEGPAMAPVALGAANTATVARAERLAISVSRRSWWQERLMLWATSGLA
Mvan_5555|M.vanbaalenii_PYR-1 LLSTGDRLPEGALAAAGTAVLDRLLRLAQPPAT-SWWRPHVAVFAVMAAC
Mflv_0897|M.gilvum_PYR-GCK ALAGHRPPVAEGLAAAATAVITRAERLVTPTRRHVRWRHRALLSVAIATM
MAB_4859|M.abscessus_ATCC_1997 DLAGTVPAAAVG--AANVAVLDRAERLLTPPEPPARAWARIALTMCVAAI
: . .. :.: * **
Mb1876c|M.bovis_AF2122/97 PTVALAVPWLTQLQRLFIA--
Rv1845c|M.tuberculosis_H37Rv PTVALAVPWLTQLQRLFIA--
MLBr_02064|M.leprae_Br4923 PTVALAVPWLTELQRLFNI--
MAV_2870|M.avium_104 PTVALAVPWLTELERLLNL--
MSMEG_3631|M.smegmatis_MC2_155 PTVAVAVPWLTELHRLFSGLT
MMAR_5059|M.marinum_M VVSPMILGLACRL--------
MUL_4135|M.ulcerans_Agy99 VVSSMFLGLACRL--------
Mvan_5555|M.vanbaalenii_PYR-1 GVPIAALSLCVY---------
Mflv_0897|M.gilvum_PYR-GCK LATPAFIQVLCHH--------
MAB_4859|M.abscessus_ATCC_1997 SAGPLVTIALATSGALLCSP-
.