For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NAALLLFAYASALIWVGPVLLRRVTGTGVHPRLAVASWLSAVFAAIAAWVGALVFLGAAVLDSIWRHRAL ALCLKELGLAGELGLPRGIASALAVGVLSTGAVATVIACWRMGRQLRRQRSASRLHAATARTIGGASDWP GVVVVPAPQPAAYCVTGRPNSVIVVTSGALGRLDEPQLAAVLAHEYAHLASRHHDLLMALRALASSLPRL PLFAAAAKVVADLLEMCADDVAVRSHGKMALLRGLLALTEGPAMAPAALGAANTATVARAERLAISVSRR SWWQERLMLWATIGLAVVSPMILGLACRL*
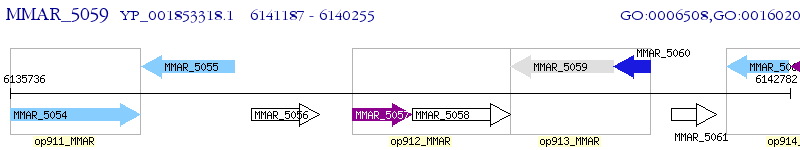
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5059 | - | - | 100% (310) | peptidase M48 like- protein |
| M. marinum M | MMAR_2719 | - | 6e-23 | 32.27% (282) | hypothetical protein MMAR_2719 |
| M. marinum M | MMAR_2906 | - | 2e-05 | 37.78% (90) | hypothetical protein MMAR_2906 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1876c | - | 2e-23 | 31.56% (282) | hypothetical protein Mb1876c |
| M. gilvum PYR-GCK | Mflv_0897 | - | 6e-65 | 44.48% (308) | hypothetical protein Mflv_0897 |
| M. tuberculosis H37Rv | Rv1845c | - | 2e-23 | 31.56% (282) | hypothetical protein Rv1845c |
| M. leprae Br4923 | MLBr_02064 | - | 2e-27 | 32.98% (282) | integral membrane protein |
| M. abscessus ATCC 19977 | MAB_4859 | - | 6e-48 | 38.96% (308) | hypothetical protein MAB_4859 |
| M. avium 104 | MAV_2870 | - | 1e-22 | 32.22% (270) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_3631 | - | 7e-20 | 29.18% (281) | integral membrane protein |
| M. thermoresistible (build 8) | TH_0125 | htpX | 6e-06 | 31.97% (122) | PROBABLE PROTEASE TRANSMEMBRANE PROTEIN HEAT SHOCK PROTEIN |
| M. ulcerans Agy99 | MUL_4135 | - | 1e-168 | 97.10% (310) | peptidase M48 like- protein |
| M. vanbaalenii PYR-1 | Mvan_3654 | - | 2e-63 | 44.48% (308) | hypothetical protein Mvan_3654 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5059|M.marinum_M MNAALLLFAYASALIWVGPVLLRRVTGTGVHPRLAVASWLSAVFAAIAAW
MUL_4135|M.ulcerans_Agy99 MNAALLLFAYASALIWVGPVLLRRVTGTGVHPRLAVASWLSAVFAAIAAW
Mflv_0897|M.gilvum_PYR-GCK MTTALWLVLYGAVLAWFAPPVLRRLTGGGVSPSMGVAAWLATVAAVLVAW
Mvan_3654|M.vanbaalenii_PYR-1 MTTALWLVLYGAVLAWLAPPVLRRLTGGGVSPSMGVAAWLATVAAVLAAW
MAB_4859|M.abscessus_ATCC_1997 MSIAVCLLLYSVAVLIFGPRLLRRLTRTGYAPRLAITAWLAAIVSVLGTW
Mb1876c|M.bovis_AF2122/97 -MSALAFTILAVLLAGPTPALLARATWPLRAPRAAMVLWQAIALAAVLSS
Rv1845c|M.tuberculosis_H37Rv -MSALAFTILAVLLAGPTPALLARATWPLRAPRAAMVLWQAIALAAVLSS
MLBr_02064|M.leprae_Br4923 -MSALAFTILAVLLVGPTPTLVARSTWPLRAPRAAMVLWQTIALAAALST
MAV_2870|M.avium_104 -MSALAFTILAVLLTGPVPAMLARARWPLRAPRAAMVLWQAVALAAVLSA
MSMEG_3631|M.smegmatis_MC2_155 -MSALAFTLVALALVGPVPAMLARASWPLRAPRAAIVLWQSIALAAVLSA
TH_0125|M.thermoresistible__bu MRRWTNWFRTAGLLGLLTALVLLVGQWVGGGTGLVYAAVVAVALNAAIYY
. : . :: . . : .
MMAR_5059|M.marinum_M VGALVFLGAAVLDSIWRHR---ALALCLKELGLAGELGLPRGIASALAVG
MUL_4135|M.ulcerans_Agy99 VGALVSLGAAVLDSIWRHR---ALALCVKELGLAGELGLPRGIASALAVG
Mflv_0897|M.gilvum_PYR-GCK LAAVVLVVDATSSSVLNGS---VVTLCLELFGFSDHTPLAGRLG---SIA
Mvan_3654|M.vanbaalenii_PYR-1 LAAVVLVVNATSNSILDGS---VVTLCLELFGFSDHTPLAGRLG---SIA
MAB_4859|M.abscessus_ATCC_1997 ISAAALIVIDVVRHWNSPA---VVLAACAARLHAMLIGQAGAAAQVGLLA
Mb1876c|M.bovis_AF2122/97 FSAGIAIASRLLMPGPDGRPTTSFVGAAGRLGWPLWAAYITVFALTVLVG
Rv1845c|M.tuberculosis_H37Rv FSAGIAIASRLLMPGPDGRPTTSFVGAAGRLGWPLWAAYITVFALTVLVG
MLBr_02064|M.leprae_Br4923 FSAGIAIASRVLMPGPDGRLTASVIGAAGRLGWPLWAAYVAAFALTVLVG
MAV_2870|M.avium_104 FSAGIAIATRVLVPGPDGRPTTSILGAEGRLGWPLWTAYIGVFALTVLVG
MSMEG_3631|M.smegmatis_MC2_155 FSAGIAIASRLFVPGADGRPTATITSEIDVLGWPLWLLYVVVFSLTLFIG
TH_0125|M.thermoresistible__bu FSDTIALRSMRARP------------------------------------
.. :
MMAR_5059|M.marinum_M VLSTGAVATVIACWRMGRQLRRQRSASRLHAATARTIGGASDWPGVVVVP
MUL_4135|M.ulcerans_Agy99 VLSTGAVATVIVCWRMGRQLRGQRSASRLHAATARTIGGVSDWPGVVVVP
Mflv_0897|M.gilvum_PYR-GCK LIGGGLLTSAVVALRLGRCLARLRTRSHEHAHAARIVGRPTDHPHVVVVE
Mvan_3654|M.vanbaalenii_PYR-1 LIGGGLLTSAVVALRLGRCLAGLRTRSHEHAHAARIVGRPTDHPHVVVVE
MAB_4859|M.abscessus_ATCC_1997 LSAAGSIAAPALGVRLARTLIRLRDTAHEHAYAVHMVGRRTAEGDVMVLE
Mb1876c|M.bovis_AF2122/97 ARLAVAVVRVATATRRRRAHHRMVVDLVGVGHNGALAQPCARARDLRVLD
Rv1845c|M.tuberculosis_H37Rv ARLAVAVVRVATATRRRRAHHRMVVDLVGVGHNGALAQPCARARDLRVLD
MLBr_02064|M.leprae_Br4923 ARLIVAIVRVAIATRRRRAHHRMVVDLVGVGHNAALAQPCARARDLRVLE
MAV_2870|M.avium_104 ARLMVAVVRVAIANRRRRAHHRMVVDLVGMGHGAALSQPCSRTRDLRVLD
MSMEG_3631|M.smegmatis_MC2_155 ARLCVAIVQVGVATRRRRAHHRMMVDLLSKSRDAVPAHLRTPASGLRILD
TH_0125|M.thermoresistible__bu -----------LSRAEAPRLHGLVAHLADQAG--------QPMPRLYISP
. : :
MMAR_5059|M.marinum_M APQPAAYCVTGRPN-SVIVVTSGALGRLDEPQLAAVLAHEYAHLASRHHD
MUL_4135|M.ulcerans_Agy99 APQPAAYCVTGRPN-SVIVVTSGALGRLDEPQLAAVLAHEYAHLASRHHD
Mflv_0897|M.gilvum_PYR-GCK ADLPAAYCVIGRP--HAIVVTSAAVRSLDRAQLKAVLAHEHAHLSGHHHH
Mvan_3654|M.vanbaalenii_PYR-1 ADLPAAYCVIGRP--HAIVVTSAAVKSLNRAQLKAVLAHENAHLTGHHHH
MAB_4859|M.abscessus_ATCC_1997 APEAAAYCVAGRP--SAIVLTTGALAVLDDAQVAAILAHERAHLAGHHPQ
Mb1876c|M.bovis_AF2122/97 VAQPLAYCLPGVR--SRVVVSEGTLTALADAEVAAILTHERAHLRARHDL
Rv1845c|M.tuberculosis_H37Rv VAQPLAYCLPGVR--SRVVVSEGTLTALADAEVAAILTHERAHLRARHDL
MLBr_02064|M.leprae_Br4923 VAQPLAYCLPGVR--SRVVVSEGALTKLNDTEVTAILTHERAHLRARHDL
MAV_2870|M.avium_104 VPQPLAYCLPGVR--SRVVVSEGTLSTLADAEVSAILTHERAHLRARHDL
MSMEG_3631|M.smegmatis_MC2_155 VAQPLAYCLPGVR--SRVVVSEGTLDTLSPDEVDAILTHERAHLRARHDL
TH_0125|M.thermoresistible__bu IRQPNAFATGRHPRHAAVCVTEGLLDILDLRELRAVLGHELAHVYNRDIL
. *:. : :: . : * :: *:* ** **: :.
MMAR_5059|M.marinum_M LLMALRALASSLPRLPLFAAAAKVVADLLEMCADDVAVRSHGKMALLRGL
MUL_4135|M.ulcerans_Agy99 LLMALRALASSLPRLPLFAAAAKVVADLLEMCADDVAVRSHGKMALLRGL
Mflv_0897|M.gilvum_PYR-GCK ILMVLRALAATLPHLPLFAGAHQAVAELLEMCADDTAARRVGTRPLLTGL
Mvan_3654|M.vanbaalenii_PYR-1 ILMVLRALAATLPHLPLFACAHQAVAELLEMCADDTAARRVGTRPLLAGL
MAB_4859|M.abscessus_ATCC_1997 LVSVLRALADVFPRVRLMTDGSAQISRLLEMCADDSAARHHGRGVLLSGL
Mb1876c|M.bovis_AF2122/97 VLEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRTPLARAL
Rv1845c|M.tuberculosis_H37Rv VLEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRTPLARAL
MLBr_02064|M.leprae_Br4923 VLEAFTAVHAAFPRLVRSSAALSAVRLLVELLADDAAVRAAGRTPLARAL
MAV_2870|M.avium_104 VLEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRAPLARAL
MSMEG_3631|M.smegmatis_MC2_155 VLEMFTAVHAAFPRFVRSASALDAVRLLIELLADDAAVRIAGPTPLARAL
TH_0125|M.thermoresistible__bu TSTVAAALAGILT----------MLADLAWFMPLGGADEEDGGGLLGRLF
*: :. : * : . . * . * * :
MMAR_5059|M.marinum_M LALTEGPAMAPAALGAANTATVARAERLAISVSRRSWWQERLMLWATIGL
MUL_4135|M.ulcerans_Agy99 LALTEGPAMAPVALGAANTATVARAERLAISVSRRSWWQERLMLWATSGL
Mflv_0897|M.gilvum_PYR-GCK LALAGHRPPVAEGLAAAATAVITRAERLVTPTRRHVRWRHRALLSVAIAT
Mvan_3654|M.vanbaalenii_PYR-1 LALAGHRPPVTEGLAAAAMAVITRAERLVTPTRRHVQWRHRALLVAAIAT
MAB_4859|M.abscessus_ATCC_1997 MDLAG--TVPAAAVGAANVAVLDRAERLLTPPEPPARAWARIALTMCVAA
Mb1876c|M.bovis_AF2122/97 VACASG-RAPSGALAVGGPSTVLRVRRLSGRGNSAVLSAAAYLAAAAVLV
Rv1845c|M.tuberculosis_H37Rv VACASG-RAPSGALAVGGPSTVLRVRRLSGRGNSAVLSAAAYLAAAAVLV
MLBr_02064|M.leprae_Br4923 VACASG-QAPSGALAAGGNTTVLRVRRLSGRSNSAVVSAAAYLAAAIVLL
MAV_2870|M.avium_104 VACASG-RAPSGALAAGGPSTVVRVRRLGGRGNSPLLSAAAYLAAAAVFV
MSMEG_3631|M.smegmatis_MC2_155 VTCAAG-RTPSGALAAGGPTTVIRVRRLGGKPNSLTLSVIAYLAAVAILV
TH_0125|M.thermoresistible__bu TLLFAPXXPAGHPVGHSATRCAPGVRGDPGRPRRRHRRRPSGGPRRRRRR
:. . ..
MMAR_5059|M.marinum_M AVVSPMILGLACRL--------
MUL_4135|M.ulcerans_Agy99 AVVSSMFLGLACRL--------
Mflv_0897|M.gilvum_PYR-GCK MLATPAFIQVLCHH--------
Mvan_3654|M.vanbaalenii_PYR-1 MLATPAFIQVLCHH--------
MAB_4859|M.abscessus_ATCC_1997 ISAGPLVTIALATSGALLCSP-
Mb1876c|M.bovis_AF2122/97 VPTVALAVPWLTQLQRLFIA--
Rv1845c|M.tuberculosis_H37Rv VPTVALAVPWLTQLQRLFIA--
MLBr_02064|M.leprae_Br4923 VPTVALAVPWLTELQRLFNI--
MAV_2870|M.avium_104 VPTVALAVPWLTELERLLNL--
MSMEG_3631|M.smegmatis_MC2_155 VPTVAVAVPWLTELHRLFSGLT
TH_0125|M.thermoresistible__bu GRRRRTTGR-------------