For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTIIFNKGTSATARIDVSAVDFGYPGMPVFRGLTLSVTPGAVTAVVGANGSGKSTLLSLLAGVLRPATGR VDVDARNIALAVQRSAVTDSFPVTAGEAVMMGRWRRLGLLRRPKADDRAVVEHWLTELGLSDLRHRALGE LSGGQRQRVLLAQAFVQQAPLLLLDEPTTGLDTDSAGVVIGHLRRLAGEGTTVVTATHDPAVVQTADHRI DLDRPGPH
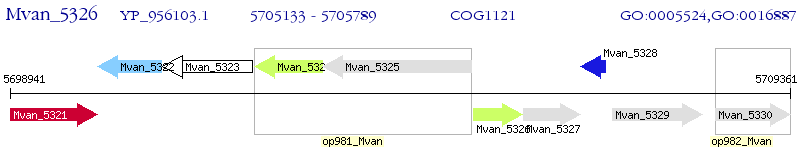
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5326 | - | - | 100% (218) | ABC transporter-related protein |
| M. vanbaalenii PYR-1 | Mvan_3370 | - | 2e-31 | 38.03% (213) | ABC transporter-related protein |
| M. vanbaalenii PYR-1 | Mvan_5319 | - | 2e-22 | 37.95% (195) | ABC transporter-related protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0074 | - | 2e-22 | 35.78% (204) | glutamine-transport ATP-binding protein ABC transporter GlnQ |
| M. gilvum PYR-GCK | Mflv_3167 | - | 2e-32 | 38.89% (216) | ABC transporter related |
| M. tuberculosis H37Rv | Rv0073 | - | 2e-22 | 35.78% (204) | glutamine-transport ATP-binding protein ABC transporter |
| M. leprae Br4923 | MLBr_00669 | ftsE | 4e-20 | 32.29% (192) | putative cell division ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_2435 | - | 3e-22 | 37.06% (197) | molybdenum ABC transporter ATP-binding protein |
| M. marinum M | MMAR_4036 | - | 2e-23 | 37.32% (209) | transmembrane ATP-binding protein ABC transporter |
| M. avium 104 | MAV_1567 | - | 4e-24 | 37.50% (216) | ABC transporter, ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_6052 | - | 1e-61 | 60.10% (198) | ABC transport protein, ATP-binding subunit |
| M. thermoresistible (build 8) | TH_4530 | - | 3e-37 | 42.13% (197) | PUTATIVE ABC transporter-related protein |
| M. ulcerans Agy99 | MUL_3901 | - | 1e-23 | 37.32% (209) | transmembrane ATP-binding protein ABC transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5326|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6052|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3167|M.gilvum_PYR-GCK --------------------------------------------------
TH_4530|M.thermoresistible__bu --------------------------------------------------
MMAR_4036|M.marinum_M MIRTWIGLVPADRRGKLILCAAVGVLSVVVRAIGTVLLVPLIGALFSSTP
MUL_3901|M.ulcerans_Agy99 MIRTWIGLVPADRRGKLILCAAVGVLSVVVRAIGTVLLVPLIGALFSSTP
MAV_1567|M.avium_104 MIRTWLSLVPADRRNKVVAYTVLALLSVAVRAVATVLLVPLVGALFSGAP
Mb0074|M.bovis_AF2122/97 --------------------------------------------------
Rv0073|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAB_2435|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5326|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6052|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3167|M.gilvum_PYR-GCK --------------------------------------------------
TH_4530|M.thermoresistible__bu --------------------------------------------------
MMAR_4036|M.marinum_M QQALVWLGWLTAATVSGWVIDAIGARVGFELGFAVLDHSQHGVADRLPGV
MUL_3901|M.ulcerans_Agy99 QQALVWLGWLTAATVSGWVIDAIGARVGFELGFAVLDHSQHGVADRLPGV
MAV_1567|M.avium_104 HRALVWLGWLTAATAIGWAIDTATARIGFNLGFAVLDHSQHDVADRLPGV
Mb0074|M.bovis_AF2122/97 --------------------------------------------------
Rv0073|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAB_2435|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5326|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6052|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3167|M.gilvum_PYR-GCK --------------------------------------------------
TH_4530|M.thermoresistible__bu --------------------------------------------------
MMAR_4036|M.marinum_M RLDWFTADNTATARQAIAASGPELVGLVVYLLTPLVSAVLLPLAIGLALL
MUL_3901|M.ulcerans_Agy99 RLDWFTADNTATARQAIAASGPELVGLVVYLLTPLVSAVLLPLAIGLALL
MAV_1567|M.avium_104 RLDWFTVEHTATARQAIAATGPELVGLVVNLLTPLISAVLLPAAIAVALV
Mb0074|M.bovis_AF2122/97 --------------------------------------------------
Rv0073|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAB_2435|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5326|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6052|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3167|M.gilvum_PYR-GCK --------------------------------------------------
TH_4530|M.thermoresistible__bu --------------------------------------------------
MMAR_4036|M.marinum_M PISWQLGLAALAGVPLLLGALWASGRFARRADKVADEANIALTERIIEFA
MUL_3901|M.ulcerans_Agy99 PISWQLGLAALAGVPLLLGALWASGRFARRADKVADEANIALTERIIEFA
MAV_1567|M.avium_104 PVSWQLGLAALGGLPLMLGALWASARLARRADSTAAEANSALTERIIEFA
Mb0074|M.bovis_AF2122/97 --------------------------------------------------
Rv0073|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAB_2435|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5326|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6052|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3167|M.gilvum_PYR-GCK --------------------------------------------------
TH_4530|M.thermoresistible__bu --------------------------------------------------
MMAR_4036|M.marinum_M RTQQALRAARRVEPARSLVGSALARQHGSLMRLFAMQVPGQLLFSIASQL
MUL_3901|M.ulcerans_Agy99 RTQQALRAARRVEPARSLVGSALARQHGSLMRLFAMQVPGQLLFSSASQL
MAV_1567|M.avium_104 RTQQALRAARRVEPARSLVGDALAAQHGATMRLLAMQVPGQLLFSLASQL
Mb0074|M.bovis_AF2122/97 --------------------------------------------------
Rv0073|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAB_2435|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5326|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6052|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3167|M.gilvum_PYR-GCK --------------------------------------------------
TH_4530|M.thermoresistible__bu --------------------------------------------------
MMAR_4036|M.marinum_M ALIVLAGATAALTVGGTLTVPEAIALIVVIARYLEPFTAVSELAPALQSV
MUL_3901|M.ulcerans_Agy99 ALIVLAGATAALTVGGTLTVPEAIALIVVIARYLEPFTAVSELAPALQSV
MAV_1567|M.avium_104 ALILLAGAATALTVNGTLTVAEAIALIVVIVRYLEPFTTISELAPALEST
Mb0074|M.bovis_AF2122/97 --------------------------------------------------
Rv0073|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAB_2435|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5326|M.vanbaalenii_PYR-1 -----------------MTIIFNKGTSATARIDVSAVDFGYP--GMPVFR
MSMEG_6052|M.smegmatis_MC2_155 ----------------------------MSAVRLNRITFGYN--RNPVFT
Mflv_3167|M.gilvum_PYR-GCK ------------------------MTTAAPALQVESLTVRYG--PVLALD
TH_4530|M.thermoresistible__bu ------------------------MT---FAIEVADVTVRYG--EVVALR
MMAR_4036|M.marinum_M RAMLDRIRSVLTAPAVAAGTATLDDGPAAARIEFDHVAFSYDGATEPVLD
MUL_3901|M.ulcerans_Agy99 RAMLDRIRSVLTAPAVAAGTATLDDGPAAARIEFDHVAFSYDGATEPVLD
MAV_1567|M.avium_104 RATLDNIRAVLTAPQMNAGSASLP-GTAAPRIEFDDVVFRYDGASVPVLD
Mb0074|M.bovis_AF2122/97 --------------------------MGDLSIQNLVVEYYSGGYALRPIN
Rv0073|M.tuberculosis_H37Rv --------------------------MGDLSIQNLVVEYYSGGYALRPIN
MLBr_00669|M.leprae_Br4923 ----------------------------MITLDHVSKKYKS--LARPALD
MAB_2435|M.abscessus_ATCC_1997 ----------------------------MSELRFDAGHLARG-------V
:
Mvan_5326|M.vanbaalenii_PYR-1 GLTLSVTPGAVTAVVGANGSGKSTLLSLLAGVLRPATGRVDVDARNIALA
MSMEG_6052|M.smegmatis_MC2_155 DLTVEFPRAACTAVTGPNGCGKSTLLSLIAGVSRPHSGTVELCARDVALA
Mflv_3167|M.gilvum_PYR-GCK DASFTLHPGRVCGLVGMNGSGKSTLFKAIMGLVRSDTGDVRIHDSSPARA
TH_4530|M.thermoresistible__bu DISARVRRGTVCGLIGMNGSGKSTLFKTIVGLVRPDTGRVLLDGDPPAVA
MMAR_4036|M.marinum_M GVSFCLQPGTTTAIVGPSGSGKSTILALIAGLHQPTGGRILIDGADAATL
MUL_3901|M.ulcerans_Agy99 GVSFCLQPGTTTAIVGPSGSGKSTILALIAGLHQPTGGRILIDGADAATL
MAV_1567|M.avium_104 GVSFSLEPGSTTAIVGPSGSGKSTILALIAGLHEPTRGRVRIDGVDAATL
Mb0074|M.bovis_AF2122/97 GLNLDVAAGSLVMLLGPSGCGKTTLLSCLGGILRPKSGAIKFDEVDITTL
Rv0073|M.tuberculosis_H37Rv GLNLDVAAGSLVMLLGPSGCGKTTLLSCLGGILRPKSGAIKFDEVDITTL
MLBr_00669|M.leprae_Br4923 NVNVKIDKGEFVFLIGPSGSGKSTFMRLLLGAETPTSGDVRVSKFHVNKL
MAB_2435|M.abscessus_ATCC_1997 RARFTVAAGETLAIVGANGAGKSTVLALVGGLLRPDTGEIRLGDHLLTNC
. . : * .*.**:*.: : * . * : .
Mvan_5326|M.vanbaalenii_PYR-1 VQR---------SAVTDSFPVTAGEAVMMG-RWRRLGLLRRPKADDRAVV
MSMEG_6052|M.smegmatis_MC2_155 VQR---------DEVARTFPITVAEAVMMG-RWRRLGLLRRPSRADRDIV
Mflv_3167|M.gilvum_PYR-GCK RRAGVVGYMPQSEAIDWTFPLSVRDVVMTG-RYGHMGPARRARHVDRRAV
TH_4530|M.thermoresistible__bu RKRGAISYVPQSEQIDWSFPLSIHDVVMTG-RYGHQGFTRRPRAADRAAV
MMAR_4036|M.marinum_M DAHTRRTLSSVVFQHPYLFHGSIRENVLAGNPGADQDQLGRAIRLARVDE
MUL_3901|M.ulcerans_Agy99 DAHTRRTLSSVVFQHPYLFHGSIRENVLAGNPGADQDQLGRAIRLARVDE
MAV_1567|M.avium_104 DAESRRAASSVVFQHPYLFHGSIRDNVLAGDPGAGDDAYARAVALARVDE
Mb0074|M.bovis_AF2122/97 QGAELANYRRNKVGIVFQAFNLVPSLTAVENVMVPLRSAGMSRRASRRRA
Rv0073|M.tuberculosis_H37Rv QGAELANYRRNKVGIVFQAFNLVPSLTAVENVMVPLRSAGMSRRASRRRA
MLBr_00669|M.leprae_Br4923 PGRHIPRLR-QVIGCVFQDFRLLQQKTVYENVAFALEVIGRRSDAINQVV
MAB_2435|M.abscessus_ATCC_1997 ATGVFVPTHRRRTALLLQQASLFPHLTVARNVEFGIRGGDVAGVRDR---
. .
Mvan_5326|M.vanbaalenii_PYR-1 EHWLTELGLSDLRHRALGELSGGQRQRVLLAQAFVQQAPLLLLDEPTTGL
MSMEG_6052|M.smegmatis_MC2_155 DFWLTELGLDDLRTCRMTELSGGQRQRTLLAQAFAQQAPILLLDEPTASL
Mflv_3167|M.gilvum_PYR-GCK DDALARVELSDFRHRQIGRLSGGQRKRAFLARCLAQGAELLLLDEPFAGV
TH_4530|M.thermoresistible__bu AAALDRVGLTDLARRQIGQASGGQRKRAFVARAIAQEADILLLDEPFAGV
MMAR_4036|M.marinum_M LIDRLPDGADTVVGEAGSALSGGERQRVSIARALLKDAPMLLVDEATSAL
MUL_3901|M.ulcerans_Agy99 LIDRLPDGADTVVGEAGSALSGGERQRVSIARALLKDAPMLLVDEATSAL
MAV_1567|M.avium_104 LVARLPDGADSVVGEAGSALSGGERQRVSIARALLKPAPILLVDEATSAL
Mb0074|M.bovis_AF2122/97 EELLARVNLAERMNHRPGDLSGGQQQRVAVARAIALDPPLILADEPTAHL
Rv0073|M.tuberculosis_H37Rv EELLARVNLAERMNHRPGDLSGGQQQRVAVARAIALDPPLILADEPTAHL
MLBr_00669|M.leprae_Br4923 PDVLETVGLSGKANRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNL
MAB_2435|M.abscessus_ATCC_1997 --WLEAVGARDLAERRPGELSGGQAQRVAIARALATEPDLVLLDEPLAGL
***: :*. :*:.: . ::* **. :
Mvan_5326|M.vanbaalenii_PYR-1 DTDSAGVVIGHLRRL-AGEGTTVVTATHDPAVVQTADHRIDLDRPGPH--
MSMEG_6052|M.smegmatis_MC2_155 DADSAQTVYRQLRRL-ASSGTTIVAATHDGDALDQFEHHLSLTGLSGHAA
Mflv_3167|M.gilvum_PYR-GCK DKRSEATICRLLRDL-ADSGATVLIATHDLNALPELADEAILLMRTVLLH
TH_4530|M.thermoresistible__bu DTRTETALIALLRQL-AADGKTVLVSTHDLRSLPTLADEVILLMRRVLAQ
MMAR_4036|M.marinum_M DTENEVAVVDALTAD-PQS-RTRVIVAHRLASIRHADRVLFVEDGRVVED
MUL_3901|M.ulcerans_Agy99 DTENEVAVVDALTAD-PQS-RTRVIVAHRLASIRHADRVLFVEDGRVVED
MAV_1567|M.avium_104 DTENEAAVVDALTAD-PRS-RTQIIVAHRLASIGHADRVLFLDHGRVVED
Mb0074|M.bovis_AF2122/97 DFIQVEEVLRLIREL-ADGERVVVVATHDSRMLP-MADRVVELTPDFAET
Rv0073|M.tuberculosis_H37Rv DFIQVEEVLRLIREL-ADGERVVVVATHDSRMLP-MADRVVELTPDFAET
MLBr_00669|M.leprae_Br4923 DPDTSKDIMDLLERI-NRTGTTVLMATHDHHIVDSMRQRVVELS------
MAB_2435|M.abscessus_ATCC_1997 DAESAPAIRALLSQVLGRDGQSALIVTHDVMDAVAVADRVLVLDSGRVAE
* : : : :*
Mvan_5326|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6052|M.smegmatis_MC2_155 TLGSGRASTCVQPQRTASRIPSDHRTP-----------------------
Mflv_3167|M.gilvum_PYR-GCK DHPDTVLDPRNLARAFGLDLPRSRGGQ-----------------------
TH_4530|M.thermoresistible__bu GDPETVLAPDNLARAFGQDVLSRR--------------------------
MMAR_4036|M.marinum_M GSIDELLAAQGRFHEFWRQQHDAAEWRIRTG-------------------
MUL_3901|M.ulcerans_Agy99 GSIDELLAAQGRFHEFWRQQHDAAEWRIRTG-------------------
MAV_1567|M.avium_104 GTVDELRAAGGRFAEFWRQQHEAAEWRIPAG-------------------
Mb0074|M.bovis_AF2122/97 NRPPETVHLQAGEVLFEQSTMGDLIYVVSEGEFEIVHDWPTAVRNWSRLP
Rv0073|M.tuberculosis_H37Rv NRPPETVHLQAGEVLFEQSTMGDLIYVVSEGEFEIVHELADGGEELVKVA
MLBr_00669|M.leprae_Br4923 --------------------LGRLVRDEWCGIYGMDR-------------
MAB_2435|M.abscessus_ATCC_1997 LGDTATVLARPTSEFGARFAGVNLVDGIAGADGALHSGDLAIHGVWAGPG
Mvan_5326|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6052|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3167|M.gilvum_PYR-GCK --------------------------------------------------
TH_4530|M.thermoresistible__bu --------------------------------------------------
MMAR_4036|M.marinum_M --------------------------------------------------
MUL_3901|M.ulcerans_Agy99 --------------------------------------------------
MAV_1567|M.avium_104 --------------------------------------------------
Mb0074|M.bovis_AF2122/97 GRG----------------ITSAR--------------------------
Rv0073|M.tuberculosis_H37Rv GPGDYFGEIGVLFHLPRSATVRARSDATAVGYTVQAFRERLGVGGLRDLI
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
MAB_2435|M.abscessus_ATCC_1997 ERVPRGQHAVAVFTPRDVAVYLSPPGGSPRNVIAVSITTLTAHGGSVVVT
Mvan_5326|M.vanbaalenii_PYR-1 -------------------------------------------
MSMEG_6052|M.smegmatis_MC2_155 -------------------------------------------
Mflv_3167|M.gilvum_PYR-GCK -------------------------------------------
TH_4530|M.thermoresistible__bu -------------------------------------------
MMAR_4036|M.marinum_M -------------------------------------------
MUL_3901|M.ulcerans_Agy99 -------------------------------------------
MAV_1567|M.avium_104 -------------------------------------------
Mb0074|M.bovis_AF2122/97 -------------------------------------------
Rv0073|M.tuberculosis_H37Rv EHRALAND-----------------------------------
MLBr_00669|M.leprae_Br4923 -------------------------------------------
MAB_2435|M.abscessus_ATCC_1997 ASRGATIFTAHITAAAAAELRLAPDTPAYFAVKAHEVVIQAAQ