For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PGPNSCPISPEFDFLDASLNLERLPVEELAELRKSEPVHWVDVPGGTGGFGDRGYWLVTKHADVKEVSKH NEIFGSSPDGAIPVWPQEMTREAIDLQKAVLLNMDAPQHTRLRKIISRGFTPRAVGRLEDELRARAQRIA ATAATEGSGDFVEQVSCELPLQAIAELLGVPQEDRDKLFRWSNEMTAGEDPEYADVDPAMSSFELITYAM KMAEERAKNPTEDIVTKLIEADIEGEKLSDDEFGFFVVMLAVAGNETTRNSITHGMIAFSRNPDQWELYK KERPETAADEIVRWATPVSAFQRTALEDTELGGVQIKKGQRVVMSYRSANFDEEVFENPHSFDIMRNPNP HVGFGGTGAHYCIGANLAKMTINLMFNAIADAMPDLKPIGDPERLKSGWLNGIKHWQVDYTGQCPVQH*
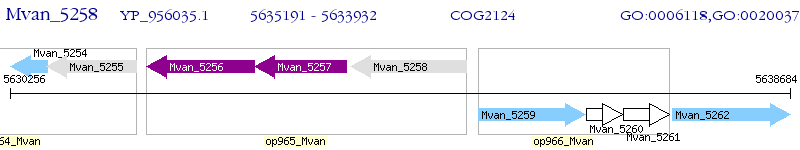
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5258 | - | - | 100% (419) | cytochrome P450 |
| M. vanbaalenii PYR-1 | Mvan_3012 | - | e-173 | 70.77% (414) | cytochrome P450 |
| M. vanbaalenii PYR-1 | Mvan_5151 | - | e-137 | 56.31% (412) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3575c | cyp125 | 0.0 | 74.29% (420) | cytochrome P450 125 |
| M. gilvum PYR-GCK | Mflv_1508 | - | 0.0 | 94.05% (420) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv3545c | cyp125 | 0.0 | 74.29% (420) | cytochrome P450 125 |
| M. leprae Br4923 | MLBr_02088 | - | 8e-39 | 30.77% (299) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_0613 | - | 1e-179 | 73.77% (408) | putative cytochrome P450 |
| M. marinum M | MMAR_5032 | cyp125A7 | 0.0 | 74.70% (415) | cytochrome P450 125A7 Cyp125A7 |
| M. avium 104 | MAV_0616 | - | 0.0 | 79.95% (419) | putative cytochrome P450 124 |
| M. smegmatis MC2 155 | MSMEG_5995 | - | 0.0 | 87.44% (430) | P450 heme-thiolate protein |
| M. thermoresistible (build 8) | TH_0362 | - | 0.0 | 84.71% (412) | PROBABLE CYTOCHROME P450 125 CYP125 |
| M. ulcerans Agy99 | MUL_4106 | cyp125A7 | 0.0 | 74.70% (415) | cytochrome P450 125A7 Cyp125A7 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3575c|M.bovis_AF2122/97 MSWNHQSVEIAVRRTTVPSPN---LPPGFDFTDPAIYAERLPVAEFAELR
Rv3545c|M.tuberculosis_H37Rv MSWNHQSVEIAVRRTTVPSPN---LPPGFDFTDPAIYAERLPVAEFAELR
MMAR_5032|M.marinum_M ----------------MPCPN---LPPGFDFTDPDIYAERLPVEEFAELR
MUL_4106|M.ulcerans_Agy99 ----------------MPCPN---LPPGFDFTDPDIYAERLPVEEFAELR
MAB_0613|M.abscessus_ATCC_1997 ----------------MVHPS---LPAGFDFTDPEIYAERLPVEELKELR
Mvan_5258|M.vanbaalenii_PYR-1 ----------------MPGPNSCPISPEFDFLDASLNLERLPVEELAELR
Mflv_1508|M.gilvum_PYR-GCK ----------------MPGTNSCPISPDFDFLDATLNLERLPVEELAELR
MSMEG_5995|M.smegmatis_MC2_155 ----------------MPTPN---IPSDFDFLDATLNLERLPVEELAELR
TH_0362|M.thermoresistible__bu ----------------MPCPN---LPKGFDPLDAELNLKGLPVAELAELR
MAV_0616|M.avium_104 ----------------MPSPN---LPPGFDLLDPDVCVKGLPVAELAELR
MLBr_02088|M.leprae_Br4923 -----MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPG
. :. . : : : .: :
Mb3575c|M.bovis_AF2122/97 SAA---PIWWNGQDPGKGGGFHDGGFWAITKLNDVKEISRHSDVFSSYEN
Rv3545c|M.tuberculosis_H37Rv SAA---PIWWNGQDPGKGGGFHDGGFWAITKLNDVKEISRHSDVFSSYEN
MMAR_5032|M.marinum_M SSE---PIWWDEQLPGQGGGFHDGGFWAITKLKDVKEVSRRSDVFSSYEN
MUL_4106|M.ulcerans_Agy99 SSE---PIWWDEQFPGQGGGFHDGGFWAITKLKDVKEVSRRSDVFSSYEN
MAB_0613|M.abscessus_ATCC_1997 KTA---PIWWQEQPDGVGG-FNDGGYWVVTKHKDVKEVSLRSDVFSSWEN
Mvan_5258|M.vanbaalenii_PYR-1 KSE---PVHWVDVPGGTGG-FGDRGYWLVTKHADVKEVSKHNEIFGSSPD
Mflv_1508|M.gilvum_PYR-GCK HSE---PVHWVDVPGGTGG-FGDKGYWLVTKHADVKEVSKRNDIFGSSPD
MSMEG_5995|M.smegmatis_MC2_155 KSE---PIHWVDVPGGTGG-FGDKGYWLVTKHADVKEVSRRSDVFGSSPD
TH_0362|M.thermoresistible__bu KSE---PIHWVDVPGGTGG-FGDKGYWIVTKHRDVKEVSRRSDVFGSSPD
MAV_0616|M.avium_104 KSA---PIYWMDVPGGTGG-FGDKGYWAITKHKDVKEISVRSDIFSSQQD
MLBr_02088|M.leprae_Br4923 TRADPFPVYRALIDYGPMQ-LPGMPLTVFSSFSDCDEALRHPLSASDRLK
*: * : . .:. * .* : .. .
Mb3575c|M.bovis_AF2122/97 GVIPRFKNDIAREDIEVQRFVMLNMDAPHHTRLRKIISRGFTPRAVGRLH
Rv3545c|M.tuberculosis_H37Rv GVIPRFKNDIAREDIEVQRFVMLNMDAPHHTRLRKIISRGFTPRAVGRLH
MMAR_5032|M.marinum_M GVIPRFKNDIAREDIDVQRFVMLNMDAPHHTRLRKIISRGFTPRAIGRLH
MUL_4106|M.ulcerans_Agy99 GVIPRFKNDIAREDIDVQRFVMLNMDAPHHTRLRKIISRGFTPRAIGRLH
MAB_0613|M.abscessus_ATCC_1997 TAIPRFQDDITREAIELQRYVMLNMDAPHHTRLRKIISRGFTPRAIGRLR
Mvan_5258|M.vanbaalenii_PYR-1 GAIPVWPQEMTREAIDLQKAVLLNMDAPQHTRLRKIISRGFTPRAVGRLE
Mflv_1508|M.gilvum_PYR-GCK GAIPVWPQDMTRDAIDLQKAVLLNMDAPQHTRLRKIISRGFTPRAVGRLE
MSMEG_5995|M.smegmatis_MC2_155 GAIPVWPQDMTREAVDLQRAVLLNMDAPQHTRLRKIISRGFTPRAIGRLE
TH_0362|M.thermoresistible__bu GAIPVWPQDMTREAIDVQRNVLLNMDAPQHTRLRKIISRGFTPRAIGRLE
MAV_0616|M.avium_104 CAIPVWPKEMTREQIDLQRNVMLNMDAPHHTRLRKIISRGFTPRAVGRLR
MLBr_02088|M.leprae_Br4923 ATLAQQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAPKVVQALE
.:. : . :: :*.*.******::*:.*:*:.: *.
Mb3575c|M.bovis_AF2122/97 DELQERAQKIAAEAAAAGSGDFVEQVSCELPLQAIAGLLGVPQEDRGKLF
Rv3545c|M.tuberculosis_H37Rv DELQERAQKIAAEAAAAGSGDFVEQVSCELPLQAIAGLLGVPQEDRGKLF
MMAR_5032|M.marinum_M DELNDRAQNIAKAAAAAGSGDFVEQVSCELPLQAIAGLLGIPQEDRGKLF
MUL_4106|M.ulcerans_Agy99 DELNDRAQNIAKAAAAAGSGDFVEQVSCELPLQAIAGLLGIPQEDRGKLF
MAB_0613|M.abscessus_ATCC_1997 DELNERAQEIAKAAAASGTGDFVEQVSCELPLQAIAGLLGVPIEDRGKLF
Mvan_5258|M.vanbaalenii_PYR-1 DELRARAQRIAATAATEGSGDFVEQVSCELPLQAIAELLGVPQEDRDKLF
Mflv_1508|M.gilvum_PYR-GCK DELRARAQRIAETAAAAGSGDFVEQVSCELPLQAIAELLGVPQDDRDKLF
MSMEG_5995|M.smegmatis_MC2_155 DELRSRAQKIAQTAAAQGAGDFVEQVSCELPLQAIAELLGVPQDDRDKLF
TH_0362|M.thermoresistible__bu DELRARAQNIAEVARSQERGDFVEQVACELPLQAIAELLGVPQDDRDKIF
MAV_0616|M.avium_104 DELDARAQNIAKTAAAAGAGDFVEQVSCELPLQAIAGLLGVPQEDRDKIF
MLBr_02088|M.leprae_Br4923 GDIAALVDSLLDKGAAAGQFDVIADLAFPLAVAVICRLLGVPYEDAPEFG
.:: .: : . : *.: ::: *.: .*. ***:* :* ::
Mb3575c|M.bovis_AF2122/97 HWSNEMTGNED--------PEYAHIDPKASSAELIGYAMKMAEEKAKNPA
Rv3545c|M.tuberculosis_H37Rv HWSNEMTGNED--------PEYAHIDPKASSAELIGYAMKMAEEKAKNPA
MMAR_5032|M.marinum_M DWSNEMTGTED--------PEFAHIDAKASSVELIGYAMKMAEEKAKNPG
MUL_4106|M.ulcerans_Agy99 DWSNEMTGTED--------PEFAHIDAKASSVELIGYAMKMAEEKAKNPG
MAB_0613|M.abscessus_ATCC_1997 NWSNEMTSYDD--------PEYADIDPAASSMEILAYSMEMAKQKAENPG
Mvan_5258|M.vanbaalenii_PYR-1 RWSNEMTAGED--------PEYADVDPAMSSFELITYAMKMAEERAKNPT
Mflv_1508|M.gilvum_PYR-GCK RWSNEMTAGED--------PEYADVDPAMSSFELITYAMKMAEERAKNPT
MSMEG_5995|M.smegmatis_MC2_155 RWSNEMTAGED--------PEYADVDPAMSSFELISYAMKMAEERAVNPT
TH_0362|M.thermoresistible__bu RWSNEMTAGED--------PEYAHIDPAMSSVELIQYAMAMADERIKNPT
MAV_0616|M.avium_104 RWSNEMTGNED--------PEYAHIDPAMSSAELIMYAMKMAEERAKNPG
MLBr_02088|M.leprae_Br4923 RVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVKCRRGTPG
* :. * * * : :. : * :.. : .*
Mb3575c|M.bovis_AF2122/97 DDIVTQLIQADIDGE-KLSDDEFGFFVVMLAVAGNETTRNSITQGMMAFA
Rv3545c|M.tuberculosis_H37Rv DDIVTQLIQADIDGE-KLSDDEFGFFVVMLAVAGNETTRNSITQGMMAFA
MMAR_5032|M.marinum_M DDIVTQLIQADIDGE-KLSDDEFGFFVVMLAVAGNETTRNSITQGMMAFA
MUL_4106|M.ulcerans_Agy99 DDIVTQLIQADIDGE-KLSDDEFGFFVVMLAVAGNETTRNSITQGMMAFA
MAB_0613|M.abscessus_ATCC_1997 EDIVTTLINAEVEGEGKLSDDEFGFFVIMLAVAGNETSRNSITQGMMAFT
Mvan_5258|M.vanbaalenii_PYR-1 EDIVTKLIEADIEGE-KLSDDEFGFFVVMLAVAGNETTRNSITHGMIAFS
Mflv_1508|M.gilvum_PYR-GCK EDIVTKLIEADIDGE-KLSDDEFGFFVVMLAVAGNETTRNSITHGMIAFS
MSMEG_5995|M.smegmatis_MC2_155 EDIVTKLIEADIDGE-KLSDDEFGFFVVMLAVAGNETTRNSITHGMIAFA
TH_0362|M.thermoresistible__bu DDIVSKLVHADIDGE-KLSEDEFGFFVVMLAVAGNETTRNSITHGMIAFS
MAV_0616|M.avium_104 DDIVTQLIQADLDGE-KLSDDEFGFFVVMLAVAGNETTRNSITHGMIAFA
MLBr_02088|M.leprae_Br4923 EDLISRLIELDESGD-QLTEEEIIATCGLLLVAGHETTVNLIANAVLAML
:*::: *:. : .*: :*:::*: :* ***:**: * *::.::*:
Mb3575c|M.bovis_AF2122/97 EHPDQWELYKK--VRPETAADEIVRWATPVTAFQRTALRDYELSGVQIKK
Rv3545c|M.tuberculosis_H37Rv EHPDQWELYKK--VRPETAADEIVRWATPVTAFQRTALRDYELSGVQIKK
MMAR_5032|M.marinum_M DNPEQWELYKR--ERPETAADEIVRWATPVTSFQRTALEDYELSGVQIKK
MUL_4106|M.ulcerans_Agy99 DNPEQWELYKR--ERPGTAADEIVRWATPVTSFQRTALEDYELSGVQIKK
MAB_0613|M.abscessus_ATCC_1997 QFPEQWELYKK--ERPETAADEIVRWATPVTSFQRTALEDTELDGVKIKK
Mvan_5258|M.vanbaalenii_PYR-1 RNPDQWELYKK--ERPETAADEIVRWATPVSAFQRTALEDTELGGVQIKK
Mflv_1508|M.gilvum_PYR-GCK QNPEQWELYKK--ERPETAADEIVRWATPVSAFQRTALEDTELGGVQIKK
MSMEG_5995|M.smegmatis_MC2_155 QNPDQWELYKK--ERPETAADEIVRWATPVSAFQRTALEDVELGGVQIKK
TH_0362|M.thermoresistible__bu QNPDQWELYKR--ERPETAADEIVRWATPVSAFQRTALEDVELSGVKIKK
MAV_0616|M.avium_104 DNPDQWELFKK--ERPETAPDEIVRWATPVTAFQRTALEDYELSGVQIKK
MLBr_02088|M.leprae_Br4923 RNPSQWKALSSNPQRAPLVVEETLRYDPAIHLIGRVAAKDMTIGQTTLTE
*.**: . *. . :* :*: ..: : *.* .* :. . :.:
Mb3575c|M.bovis_AF2122/97 GQRVVMFYRSANFDEEVFQDPFTFNILRNPNPHVGFGGTGAHYCIGANLA
Rv3545c|M.tuberculosis_H37Rv GQRVVMFYRSANFDEEVFQDPFTFNILRNPNPHVGFGGTGAHYCIGANLA
MMAR_5032|M.marinum_M GQRVLMFYRSANFDEEVFEDPFSFNILRNPNPHVGFGGTGAHYCIGANLA
MUL_4106|M.ulcerans_Agy99 GQRVLMFYRSANFDEEVFEDPFSFNILRNPNPHVGFGGTGAHYCIGANLA
MAB_0613|M.abscessus_ATCC_1997 GQRVVMMYRSANFDEDVFEDPFSFNIMRNPNPHMGFGGSGAHYCIGANLA
Mvan_5258|M.vanbaalenii_PYR-1 GQRVVMSYRSANFDEEVFENPHSFDIMRNPNPHVGFGGTGAHYCIGANLA
Mflv_1508|M.gilvum_PYR-GCK GQRVVMSYRSANFDDEVFDNPHSFDITRNPNPHVGFGGTGAHYCIGANLA
MSMEG_5995|M.smegmatis_MC2_155 GQRVVMSYRSANFDEEVFEDPHTFNILRSPNPHVGFGGTGAHYCIGANLA
TH_0362|M.thermoresistible__bu GERVVMSYRSANFDEEVFEDPHTFNILRDPNPHVGFGGTGAHYCIGANLA
MAV_0616|M.avium_104 GQRVVMFYRSANFDEEVFEDPHRFNILRNPNPHVGFGGTGAHYCIGANLA
MLBr_02088|M.leprae_Br4923 GDTMVLLLAAANRDPAVYSRPDEFDPDRPSSRHLAFA-VGSHFCLGAALA
*: ::: :** * *:. * *: * .. *:.*. *:*:*:** **
Mb3575c|M.bovis_AF2122/97 RMTINLIFNAVADHMPDLKPISAPERLRSGWLNGIKHWQVDYTGR-----
Rv3545c|M.tuberculosis_H37Rv RMTINLIFNAVADHMPDLKPISAPERLRSGWLNGIKHWQVDYTGR-----
MMAR_5032|M.marinum_M RMTINLIFNAVADHMPDLTPIAAPERLRSGWLNGIKHWQVDYTGK-----
MUL_4106|M.ulcerans_Agy99 RMTINLIFNAVADHMPDLKPIAAPERLRSGWLNGIKHWQVDYTGK-----
MAB_0613|M.abscessus_ATCC_1997 RLTINLMFNAIADHMPNLAPAGDPKRLQSGWLNGIKHWQVDFTGAS----
Mvan_5258|M.vanbaalenii_PYR-1 KMTINLMFNAIADAMPDLKPIGDPERLKSGWLNGIKHWQVDYTG------
Mflv_1508|M.gilvum_PYR-GCK KMTINLIFNAIADAMPDLKPIGEPERLKSGWLNGIKHWQVDYSGK-----
MSMEG_5995|M.smegmatis_MC2_155 RMTINLIFNAIADNMPDLKPIGAPERLKSGWLNGIKHWQVDYTGAGKASV
TH_0362|M.thermoresistible__bu KLTINLMFNAIADHMPDLTPIGTPERLKSGWLNGIKHWQVDYTG------
MAV_0616|M.avium_104 RMTISLIFNAVADHMPDLKPLSAPERLRSGWLNGIKHWQVDYTG------
MLBr_02088|M.leprae_Br4923 RLEATVTLSAISARFPQVQLAGELVYKPNVAMRGMSALPVQV--------
:: .: :.*:: :*:: . . :.*:. *:
Mb3575c|M.bovis_AF2122/97 ------CPVAH
Rv3545c|M.tuberculosis_H37Rv ------CPVAH
MMAR_5032|M.marinum_M ------CPVSH
MUL_4106|M.ulcerans_Agy99 ------CPVSH
MAB_0613|M.abscessus_ATCC_1997 -----GCPVLQ
Mvan_5258|M.vanbaalenii_PYR-1 -----QCPVQH
Mflv_1508|M.gilvum_PYR-GCK -----GCPVSH
MSMEG_5995|M.smegmatis_MC2_155 SGAPGTCPVAH
TH_0362|M.thermoresistible__bu -----TCPVAK
MAV_0616|M.avium_104 -----KCPVAH
MLBr_02088|M.leprae_Br4923 -----------