For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVHPSLPAGFDFTDPEIYAERLPVEELKELRKTAPIWWQEQPDGVGGFNDGGYWVVTKHKDVKEVSLRSD VFSSWENTAIPRFQDDITREAIELQRYVMLNMDAPHHTRLRKIISRGFTPRAIGRLRDELNERAQEIAKA AAASGTGDFVEQVSCELPLQAIAGLLGVPIEDRGKLFNWSNEMTSYDDPEYADIDPAASSMEILAYSMEM AKQKAENPGEDIVTTLINAEVEGEGKLSDDEFGFFVIMLAVAGNETSRNSITQGMMAFTQFPEQWELYKK ERPETAADEIVRWATPVTSFQRTALEDTELDGVKIKKGQRVVMMYRSANFDEDVFEDPFSFNIMRNPNPH MGFGGSGAHYCIGANLARLTINLMFNAIADHMPNLAPAGDPKRLQSGWLNGIKHWQVDFTGASGCPVLQ
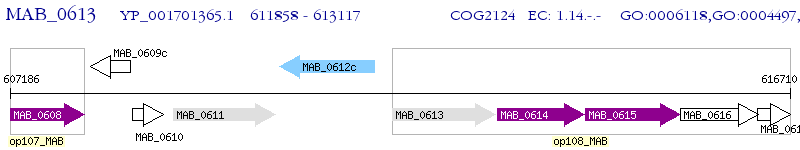
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0613 | - | - | 100% (419) | putative cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_0611 | - | e-170 | 69.76% (410) | putative cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_1211c | - | e-136 | 54.85% (412) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3575c | cyp125 | 0.0 | 76.32% (418) | cytochrome P450 125 |
| M. gilvum PYR-GCK | Mflv_3290 | - | 0.0 | 72.71% (414) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv3545c | cyp125 | 0.0 | 76.32% (418) | cytochrome P450 125 |
| M. leprae Br4923 | MLBr_02088 | - | 1e-32 | 28.43% (299) | putative cytochrome p450 |
| M. marinum M | MMAR_5032 | cyp125A7 | 0.0 | 78.55% (415) | cytochrome P450 125A7 Cyp125A7 |
| M. avium 104 | MAV_2811 | - | 0.0 | 73.80% (416) | putative cytochrome P450 124 |
| M. smegmatis MC2 155 | MSMEG_5995 | - | 0.0 | 73.68% (418) | P450 heme-thiolate protein |
| M. thermoresistible (build 8) | TH_1956 | - | 1e-178 | 70.02% (417) | PROBABLE CYTOCHROME P450 125 CYP125 |
| M. ulcerans Agy99 | MUL_4106 | cyp125A7 | 0.0 | 78.31% (415) | cytochrome P450 125A7 Cyp125A7 |
| M. vanbaalenii PYR-1 | Mvan_3012 | - | 1e-179 | 71.33% (415) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3290|M.gilvum_PYR-GCK ----------------MATPTLPPGFDFTDPDLNRERLPVEELAELRRCA
Mvan_3012|M.vanbaalenii_PYR-1 ----------------MATPTLPPGFDFTDPDLNLERLPVEELAELRRCA
MAV_2811|M.avium_104 --------MATVEPTTKPVPNLPPGFDFTDPDIYAERLPVEELAEMRRVA
TH_1956|M.thermoresistible__bu ----------------MPTPNIPPGFDFTDPDIYAVRLPVEELAELRRSA
Mb3575c|M.bovis_AF2122/97 MSWNHQSVEIAVRRTTVPSPNLPPGFDFTDPAIYAERLPVAEFAELRSAA
Rv3545c|M.tuberculosis_H37Rv MSWNHQSVEIAVRRTTVPSPNLPPGFDFTDPAIYAERLPVAEFAELRSAA
MMAR_5032|M.marinum_M ----------------MPCPNLPPGFDFTDPDIYAERLPVEEFAELRSSE
MUL_4106|M.ulcerans_Agy99 ----------------MPCPNLPPGFDFTDPDIYAERLPVEEFAELRSSE
MAB_0613|M.abscessus_ATCC_1997 ----------------MVHPSLPAGFDFTDPEIYAERLPVEELKELRKTA
MSMEG_5995|M.smegmatis_MC2_155 ----------------MPTPNIPSDFDFLDATLNLERLPVEELAELRKSE
MLBr_02088|M.leprae_Br4923 --MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRA
. :. . : :: : .: :
Mflv_3290|M.gilvum_PYR-GCK PIWWNEQPDG-IGGFGDGGF--WVVTKHHDVKEISKRSDVFSSEVKTALP
Mvan_3012|M.vanbaalenii_PYR-1 PIWWNEQTSGGAGPFGDGGY--WVVTKHRDVKEISKHSEVFSSQQKTALP
MAV_2811|M.avium_104 PIWWNEQPIG-AGGFDDGGF--WVVTKHKDVKEVSLRSDVFSSLQKTALP
TH_1956|M.thermoresistible__bu PIWWNEQPMG-AGGFDDGGF--WLVTRHSDVKEVSRRSDVFSSQRKTALP
Mb3575c|M.bovis_AF2122/97 PIWWNGQDPGKGGGFHDGGF--WAITKLNDVKEISRHSDVFSSYENGVIP
Rv3545c|M.tuberculosis_H37Rv PIWWNGQDPGKGGGFHDGGF--WAITKLNDVKEISRHSDVFSSYENGVIP
MMAR_5032|M.marinum_M PIWWDEQLPGQGGGFHDGGF--WAITKLKDVKEVSRRSDVFSSYENGVIP
MUL_4106|M.ulcerans_Agy99 PIWWDEQFPGQGGGFHDGGF--WAITKLKDVKEVSRRSDVFSSYENGVIP
MAB_0613|M.abscessus_ATCC_1997 PIWWQEQPDGVGG-FNDGGY--WVVTKHKDVKEVSLRSDVFSSWENTAIP
MSMEG_5995|M.smegmatis_MC2_155 PIHWVDVPGGTGG-FGDKGY--WLVTKHADVKEVSRRSDVFGSSPDGAIP
MLBr_02088|M.leprae_Br4923 DPFPVYRALIDYGPMQLPGMPLTVFSSFSDCDEALRHPLSASDRLKATLA
* : * .: * .* :. .. . .:.
Mflv_3290|M.gilvum_PYR-GCK RYPEGSTGEQIETGSLVLLNMDAPRHTHLRKIISRGFTPRAVERLRDDLN
Mvan_3012|M.vanbaalenii_PYR-1 RYPEGSTTEQVETGSLVLLNMDAPRHTHLRKIISRGFTPRAVERLREDLA
MAV_2811|M.avium_104 RYKDGTVAEQVERGKFVLLNMDAPQHTRLRKIISRAFTPRAVERLRDDLR
TH_1956|M.thermoresistible__bu RYRDGTVAEQIDRGKYVLLNMDAPHHTRLRKIISRAFTPRAVERLRDELN
Mb3575c|M.bovis_AF2122/97 RFKNDIAREDIEVQRFVMLNMDAPHHTRLRKIISRGFTPRAVGRLHDELQ
Rv3545c|M.tuberculosis_H37Rv RFKNDIAREDIEVQRFVMLNMDAPHHTRLRKIISRGFTPRAVGRLHDELQ
MMAR_5032|M.marinum_M RFKNDIAREDIDVQRFVMLNMDAPHHTRLRKIISRGFTPRAIGRLHDELN
MUL_4106|M.ulcerans_Agy99 RFKNDIAREDIDVQRFVMLNMDAPHHTRLRKIISRGFTPRAIGRLHDELN
MAB_0613|M.abscessus_ATCC_1997 RFQDDITREAIELQRYVMLNMDAPHHTRLRKIISRGFTPRAIGRLRDELN
MSMEG_5995|M.smegmatis_MC2_155 VWPQDMTREAVDLQRAVLLNMDAPQHTRLRKIISRGFTPRAIGRLEDELR
MLBr_02088|M.leprae_Br4923 QQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAPKVVQALEGDIA
* :: :*.* **:***::*:.*:*:.: *. ::
Mflv_3290|M.gilvum_PYR-GCK ARAQNIAKTAASAGSGDFVEQVSCELPLQAIAGLLGVPVDDRKKLFDWSN
Mvan_3012|M.vanbaalenii_PYR-1 QRAHNIAKSAAAAGAGDFVEQVSCELPLQAIAGLLGVPLEDRKKLFDWSN
MAV_2811|M.avium_104 ERARRIVEAAAAEGSGDFVEQVSCELPLQAIASLMGVPQEDRKKLFHWSN
TH_1956|M.thermoresistible__bu ERAHRIVMEAAAAGSGDFVEQVACELPLQAIAGLMGVPQEDRKKLFDWSN
Mb3575c|M.bovis_AF2122/97 ERAQKIAAEAAAAGSGDFVEQVSCELPLQAIAGLLGVPQEDRGKLFHWSN
Rv3545c|M.tuberculosis_H37Rv ERAQKIAAEAAAAGSGDFVEQVSCELPLQAIAGLLGVPQEDRGKLFHWSN
MMAR_5032|M.marinum_M DRAQNIAKAAAAAGSGDFVEQVSCELPLQAIAGLLGIPQEDRGKLFDWSN
MUL_4106|M.ulcerans_Agy99 DRAQNIAKAAAAAGSGDFVEQVSCELPLQAIAGLLGIPQEDRGKLFDWSN
MAB_0613|M.abscessus_ATCC_1997 ERAQEIAKAAAASGTGDFVEQVSCELPLQAIAGLLGVPIEDRGKLFNWSN
MSMEG_5995|M.smegmatis_MC2_155 SRAQKIAQTAAAQGAGDFVEQVSCELPLQAIAELLGVPQDDRDKLFRWSN
MLBr_02088|M.leprae_Br4923 ALVDSLLDKGAAAGQFDVIADLAFPLAVAVICRLLGVPYEDAPEFGRVSA
. : .*: * *.: ::: *.: .*. *:*:* :* :: *
Mflv_3290|M.gilvum_PYR-GCK QMVSDDD--------PEYAHYDNRNAATELIMYAMQLAAVRAEQPGEDIV
Mvan_3012|M.vanbaalenii_PYR-1 QMVSDDD--------PEFAHYDNRNAATELIMYAMQLAALRAEQPGEDIV
MAV_2811|M.avium_104 EMVGDQD--------PEFASNDAITASVELIMYGMQMAADRAKNPGEDLV
TH_1956|M.thermoresistible__bu QMVGDQD--------PEFADNDAIGASIELIGYGMQLAADRQRNPRDDLV
Mb3575c|M.bovis_AF2122/97 EMTGNED--------PEYAHIDPKASSAELIGYAMKMAEEKAKNPADDIV
Rv3545c|M.tuberculosis_H37Rv EMTGNED--------PEYAHIDPKASSAELIGYAMKMAEEKAKNPADDIV
MMAR_5032|M.marinum_M EMTGTED--------PEFAHIDAKASSVELIGYAMKMAEEKAKNPGDDIV
MUL_4106|M.ulcerans_Agy99 EMTGTED--------PEFAHIDAKASSVELIGYAMKMAEEKAKNPGDDIV
MAB_0613|M.abscessus_ATCC_1997 EMTSYDD--------PEYADIDPAASSMEILAYSMEMAKQKAENPGEDIV
MSMEG_5995|M.smegmatis_MC2_155 EMTAGED--------PEYADVDPAMSSFELISYAMKMAEERAVNPTEDIV
MLBr_02088|M.leprae_Br4923 LLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVKCRRGTPGEDLI
:. * * * : :. : * ::. : * :*::
Mflv_3290|M.gilvum_PYR-GCK TKLIEADVDGH-KLSDDEFGFFMVLLAVAGNETTRNSITHGMIAFTEHPD
Mvan_3012|M.vanbaalenii_PYR-1 TKLIEADVDGH-KLTDDEFGFFMVLLAVAGNETTRNSITHGMIAFTEHPD
MAV_2811|M.avium_104 TKLVQADIDGH-KLSDDEFGFFVILLAVAGNETTRNSITQGMMAFTDFPD
TH_1956|M.thermoresistible__bu TTLVQADVDGH-KLSDDEFGFFVILLAVAGNETTRNSITQGMMAFTEHPD
Mb3575c|M.bovis_AF2122/97 TQLIQADIDGE-KLSDDEFGFFVVMLAVAGNETTRNSITQGMMAFAEHPD
Rv3545c|M.tuberculosis_H37Rv TQLIQADIDGE-KLSDDEFGFFVVMLAVAGNETTRNSITQGMMAFAEHPD
MMAR_5032|M.marinum_M TQLIQADIDGE-KLSDDEFGFFVVMLAVAGNETTRNSITQGMMAFADNPE
MUL_4106|M.ulcerans_Agy99 TQLIQADIDGE-KLSDDEFGFFVVMLAVAGNETTRNSITQGMMAFADNPE
MAB_0613|M.abscessus_ATCC_1997 TTLINAEVEGEGKLSDDEFGFFVIMLAVAGNETSRNSITQGMMAFTQFPE
MSMEG_5995|M.smegmatis_MC2_155 TKLIEADIDGE-KLSDDEFGFFVVMLAVAGNETTRNSITHGMIAFAQNPD
MLBr_02088|M.leprae_Br4923 SRLIELDESGD-QLTEEEIIATCGLLLVAGHETTVNLIANAVLAMLRNPS
: *:: : .*. :*:::*: :* ***:**: * *::.::*: *.
Mflv_3290|M.gilvum_PYR-GCK QWELYKR--ERPITAVDEIVRWATPVTSFQRTALTDYELSGVQITKGQRV
Mvan_3012|M.vanbaalenii_PYR-1 QWELFKR--ERPATAVDEIVRWATPVTSFQRTALRDYELSGVQIKKGQRV
MAV_2811|M.avium_104 QWELFKR--ERPATAADEIVRWATPVTSFQRTALQDYELSGVKIKKGQRV
TH_1956|M.thermoresistible__bu QWELFKE--QRPPTTADEIVRWATPVTSFQRTALADVELSGVQIKAGQRV
Mb3575c|M.bovis_AF2122/97 QWELYKK--VRPETAADEIVRWATPVTAFQRTALRDYELSGVQIKKGQRV
Rv3545c|M.tuberculosis_H37Rv QWELYKK--VRPETAADEIVRWATPVTAFQRTALRDYELSGVQIKKGQRV
MMAR_5032|M.marinum_M QWELYKR--ERPETAADEIVRWATPVTSFQRTALEDYELSGVQIKKGQRV
MUL_4106|M.ulcerans_Agy99 QWELYKR--ERPGTAADEIVRWATPVTSFQRTALEDYELSGVQIKKGQRV
MAB_0613|M.abscessus_ATCC_1997 QWELYKK--ERPETAADEIVRWATPVTSFQRTALEDTELDGVKIKKGQRV
MSMEG_5995|M.smegmatis_MC2_155 QWELYKK--ERPETAADEIVRWATPVSAFQRTALEDVELGGVQIKKGQRV
MLBr_02088|M.leprae_Br4923 QWKALSSNPQRAPLVVEETLRYDPAIHLIGRVAAKDMTIGQTTLTEGDTM
**: . *. ..:* :*: ..: : *.* * :. . :. *: :
Mflv_3290|M.gilvum_PYR-GCK VMSYRSANFDEEVFDDPFTFDILRDPNPHVGFGGTGAHYCIGANLARMTI
Mvan_3012|M.vanbaalenii_PYR-1 VMSYRSANFDEEVFDDPFTFDIMRDPNPHVGFGGTGAHYCIGANLARMTI
MAV_2811|M.avium_104 VMFYRSANFDEDVFDDPFTFNILRDPNPHVGFGGTGAHYCIGANLARMTI
TH_1956|M.thermoresistible__bu VMCYRSANFDDTVFDDPYTFNILRDPNPHLGFGGTGAHYCIGANLARLTI
Mb3575c|M.bovis_AF2122/97 VMFYRSANFDEEVFQDPFTFNILRNPNPHVGFGGTGAHYCIGANLARMTI
Rv3545c|M.tuberculosis_H37Rv VMFYRSANFDEEVFQDPFTFNILRNPNPHVGFGGTGAHYCIGANLARMTI
MMAR_5032|M.marinum_M LMFYRSANFDEEVFEDPFSFNILRNPNPHVGFGGTGAHYCIGANLARMTI
MUL_4106|M.ulcerans_Agy99 LMFYRSANFDEEVFEDPFSFNILRNPNPHVGFGGTGAHYCIGANLARMTI
MAB_0613|M.abscessus_ATCC_1997 VMMYRSANFDEDVFEDPFSFNIMRNPNPHMGFGGSGAHYCIGANLARLTI
MSMEG_5995|M.smegmatis_MC2_155 VMSYRSANFDEEVFEDPHTFNILRSPNPHVGFGGTGAHYCIGANLARMTI
MLBr_02088|M.leprae_Br4923 VLLLAAANRDPAVYSRPDEFDPDRPSSRHLAFA-VGSHFCLGAALARLEA
:: :** * *:. * *: * .. *:.*. *:*:*:** ***:
Mflv_3290|M.gilvum_PYR-GCK DLMFNAIADHMPDLSAIGSPDRLRSGWLNGIKHWQVDYSG----------
Mvan_3012|M.vanbaalenii_PYR-1 DLMFNAIADHLPDLSSAGTPDRLRSGWLNGIKHWQVDYTGP---------
MAV_2811|M.avium_104 DLMFNAIADAMPDLESIGKPERLRSGWLNGIKHWQVDYHTNGS-------
TH_1956|M.thermoresistible__bu DLIFAAIAEHLPDLRPLGPPERLRSGWLNGIKHWPVDYGAPRV-------
Mb3575c|M.bovis_AF2122/97 NLIFNAVADHMPDLKPISAPERLRSGWLNGIKHWQVDYTGR---------
Rv3545c|M.tuberculosis_H37Rv NLIFNAVADHMPDLKPISAPERLRSGWLNGIKHWQVDYTGR---------
MMAR_5032|M.marinum_M NLIFNAVADHMPDLTPIAAPERLRSGWLNGIKHWQVDYTGK---------
MUL_4106|M.ulcerans_Agy99 NLIFNAVADHMPDLKPIAAPERLRSGWLNGIKHWQVDYTGK---------
MAB_0613|M.abscessus_ATCC_1997 NLMFNAIADHMPNLAPAGDPKRLQSGWLNGIKHWQVDFTGAS--------
MSMEG_5995|M.smegmatis_MC2_155 NLIFNAIADNMPDLKPIGAPERLKSGWLNGIKHWQVDYTGAGKASVSGAP
MLBr_02088|M.leprae_Br4923 TVTLSAISARFPQVQLAGELVYKPNVAMRGMSALPVQV------------
: : *:: :*:: . . :.*:. *:
Mflv_3290|M.gilvum_PYR-GCK RGCPVAH----
Mvan_3012|M.vanbaalenii_PYR-1 SGCPVAH----
MAV_2811|M.avium_104 SKCPVAH----
TH_1956|M.thermoresistible__bu PTGAAGPQRCG
Mb3575c|M.bovis_AF2122/97 --CPVAH----
Rv3545c|M.tuberculosis_H37Rv --CPVAH----
MMAR_5032|M.marinum_M --CPVSH----
MUL_4106|M.ulcerans_Agy99 --CPVSH----
MAB_0613|M.abscessus_ATCC_1997 -GCPVLQ----
MSMEG_5995|M.smegmatis_MC2_155 GTCPVAH----
MLBr_02088|M.leprae_Br4923 -----------