For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGAAAYVGRVGGLAVALGVGTAVFAGQGVAWADDTESSTNGASSAATSDSVQTDTTSDTVDTPEADTEDE TEPDPEPEDQAEPDPEPEDQAEPDPEPEPEPQDEPDPDQDQDQDADDSEGADADQEPPKATETNTETSTK PEPEPEPEPEPTEKTTATVTDDGSDDPAVPPQEPKKVAVFATATSLAESSPKTLSPKVAALLTTEQDEAA AAKKPNLITAVVNVVTSMLDWARQKADAKPGDAPQPPFLWALLSFARRELDNLFAARSTTNAPRTTSVTP TSLALTDDATTAALIPSYSPWLNPQVSPSTRFVSWVTGKYVYLDDDTLANTLARFSVYGTDVGVMWDNGM VDDPSTPDINEHQILIAVGDTFGAKGMTGRHIYNTLFRSSDTDLSDGMTIPDGEWFNGNMFGGAPLDGPT QARPIINRPSWLPNSVTLIPTAGVSLPTEVTEDTPFGTIQYVSFMSVSNWGSSGRWTTNYSAIAYSTDNG ENFKVAPESVRYNSIFSGNRNFQQSAFVKGDDGYVYVYGTPNGRQGAAYVARVNNENILDVSEYEYYKAA STGWFGSSEARWVKGNPSSASAVIGKTGGACGSTEPGYTVSEMSVQYNEYLQKYIVLYGDQFNNIVMRTS DTPEGAWSDATVLMGQQSGGIYAPMLHPWSPSTQGTGSDLYWNLSLWSDYNVMLMKTDLTKL
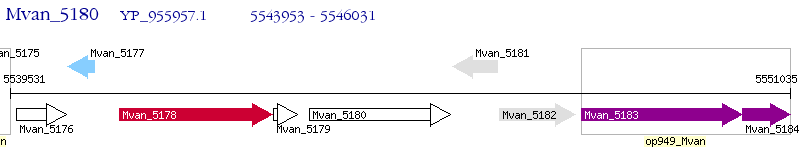
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5180 | - | - | 100% (692) | hypothetical protein Mvan_5180 |
| M. vanbaalenii PYR-1 | Mvan_5030 | - | 4e-29 | 34.27% (286) | glycoside hydrolase family protein |
| M. vanbaalenii PYR-1 | Mvan_1255 | - | 2e-23 | 31.91% (257) | hypothetical protein Mvan_1255 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1783c | - | 2e-82 | 36.72% (542) | hypothetical protein Mb1783c |
| M. gilvum PYR-GCK | Mflv_1576 | - | 0.0 | 66.52% (684) | hypothetical protein Mflv_1576 |
| M. tuberculosis H37Rv | Rv1754c | - | 2e-82 | 36.72% (542) | hypothetical protein Rv1754c |
| M. leprae Br4923 | MLBr_02491 | - | 1e-73 | 41.51% (371) | hypothetical protein MLBr_02491 |
| M. abscessus ATCC 19977 | MAB_3454 | - | 3e-83 | 40.74% (405) | hypothetical protein MAB_3454 |
| M. marinum M | MMAR_2586 | - | 7e-85 | 38.71% (527) | hypothetical protein MMAR_2586 |
| M. avium 104 | MAV_1035 | - | 2e-79 | 35.59% (562) | hypothetical protein MAV_1035 |
| M. smegmatis MC2 155 | MSMEG_4365 | - | 7e-87 | 31.90% (765) | hypothetical protein MSMEG_4365 |
| M. thermoresistible (build 8) | TH_1246 | - | 2e-74 | 41.58% (392) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3174 | - | 5e-85 | 38.71% (527) | hypothetical protein MUL_3174 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1783c|M.bovis_AF2122/97 --------------------------------------------------
Rv1754c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2586|M.marinum_M --------------------------------------------------
MUL_3174|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02491|M.leprae_Br4923 --------------------------------------------------
MAV_1035|M.avium_104 --------------------------------------------------
MAB_3454|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5180|M.vanbaalenii_PYR-1 MGAAAYVGRVGGLAVALGVGTAVFAGQGVAWADDTESSTNGASSAATS--
Mflv_1576|M.gilvum_PYR-GCK ----------------------------MAWADDGASSASKPSASATSGG
TH_1246|M.thermoresistible__bu --------------------------------------------------
MSMEG_4365|M.smegmatis_MC2_155 ------MGRIGALAVALGIGAALTTGTGVAWADDSGASGGSTS-------
Mb1783c|M.bovis_AF2122/97 -------------------------MYRYQVRVQQRRSEMNRWVATRSRR
Rv1754c|M.tuberculosis_H37Rv -------------------------MYRYQVRVQQRRSEMNRWVATRSRR
MMAR_2586|M.marinum_M --------------------------------------------------
MUL_3174|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02491|M.leprae_Br4923 --------------------------------------------------
MAV_1035|M.avium_104 --------------------------------------------------
MAB_3454|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5180|M.vanbaalenii_PYR-1 ---DSVQTDTTSDTVDTPEADTEDETEPDPEPEDQAEPDPEPEDQAEPDP
Mflv_1576|M.gilvum_PYR-GCK NTADTQSTTTKPDDATQTDDDTADDTATDDSATDTEELGAEDPDVEEPDI
TH_1246|M.thermoresistible__bu --------------------------------------------------
MSMEG_4365|M.smegmatis_MC2_155 ----SSSSASDSSSSSAGGSSGSASGGADAAKPRKPGVTKPRIQPTGKIA
Mb1783c|M.bovis_AF2122/97 HTYQWITDHKSPRDHYRHISELRTSIATSSPGRCDMSPIPRIVSVSLAWA
Rv1754c|M.tuberculosis_H37Rv HTYQWITDHKSPRDHYRHISELRTSIATSSPGRCDMSPIPRIVSVSLAWA
MMAR_2586|M.marinum_M -----------------------------------MSTIHRLVSLSVASA
MUL_3174|M.ulcerans_Agy99 -----------------------------------MSTIHRLVSLSVASA
MLBr_02491|M.leprae_Br4923 --------------------------------------------------
MAV_1035|M.avium_104 ---------------------MDVNAVSNPLERCYMSAFLRSVSLSMASA
MAB_3454|M.abscessus_ATCC_1997 -----------------------------------MSTR-RRISVVLMTS
Mvan_5180|M.vanbaalenii_PYR-1 EPEPEPQDEPDPDQD-QDQDADDSEGADADQEPPKATETNTETSTKPEPE
Mflv_1576|M.gilvum_PYR-GCK DSPQVDTDSPDDEVDGPDDEAEPEPAAESDDRTPAKPAGRAERDTEPEAE
TH_1246|M.thermoresistible__bu --------------------------------------------------
MSMEG_4365|M.smegmatis_MC2_155 ESLKKTAKAVEGAVADAHKRAQQSAAQQAEKRAERRAKLRAQIRPGKDTD
Mb1783c|M.bovis_AF2122/97 AAIGLMVPIGLAPPAMAAPCSGDAANAPPPPSAIVTDPGA----TALGPV
Rv1754c|M.tuberculosis_H37Rv AAIGLMVPIGLAPPAMAAPCSGDAANAPPPPSAIVTDPGA----TALGPV
MMAR_2586|M.marinum_M AAFGFIGAVGLAPMASAVPCSGDAANAPPPANAVVTAPSS----KTLG--
MUL_3174|M.ulcerans_Agy99 AAFGFIGAVGLAPMASAVPCSGDAANAPPPANAVVTAPSS----KTLG--
MLBr_02491|M.leprae_Br4923 --------------------------------------------------
MAV_1035|M.avium_104 AAVGFVLVVGPAPTANATPCGAPEANVDPPAAPPAMPAPQPVVQP-----
MAB_3454|M.abscessus_ATCC_1997 AVIGSSVAVDVAPVAYADPCAGPAAGLQPPTPVPDDGIPG----------
Mvan_5180|M.vanbaalenii_PYR-1 PEPEPEPTEKTTATVTDDGSDDP--AVPPQEPKKVAVFATATSLAESSPK
Mflv_1576|M.gilvum_PYR-GCK VEAGPQQTAQTSERDPSDTTATPRDTEDIPSPQTSTTVTVDATAAKATTG
TH_1246|M.thermoresistible__bu --------------------------------------------------
MSMEG_4365|M.smegmatis_MC2_155 KQAPEKQAGNEAKNAAQPATKTQRQLPAAPAVKVPDVITSVRARSDEVAR
Mb1783c|M.bovis_AF2122/97 RPGHGPIPTGRKPRGANDRAPLPKLGPLISALLN---PGARNAAPLQQQA
Rv1754c|M.tuberculosis_H37Rv RPGHGPIPTGRKPRGANDRAPLPKLGPLISALLN---PGARNAAPLQQQA
MMAR_2586|M.marinum_M -----PMTRGLRPRGANERAPLPKLGP-LSRLLN---PGARRSAPVQQQA
MUL_3174|M.ulcerans_Agy99 -----PMTRGLRPRGANERAPLPKLGP-LSRLLN---PGARRSAPVQQQA
MLBr_02491|M.leprae_Br4923 --------------------------------------------------
MAV_1035|M.avium_104 -------PTGRRPTHTNDQAPLPKLGPLISSLLKPSAPGRQYSAPVHPRA
MAB_3454|M.abscessus_ATCC_1997 ----QAPPIGRRPAGANDKAPLPELGKLPLAILKQVLPQQSAKKKPGWAQ
Mvan_5180|M.vanbaalenii_PYR-1 TLSPKVAALLTTEQDEAAAAKKPNLITAVVNVVTSMLDWARQKADAKPGD
Mflv_1576|M.gilvum_PYR-GCK TASSTLADLSTTEQGPQTADAKPNLITAVVNVVNSMIDWARQKAAADPGD
TH_1246|M.thermoresistible__bu --------------------------------------------------
MSMEG_4365|M.smegmatis_MC2_155 EITTGVTGIRTAIENAAPRIALPDLRP-TTNLARPVTPVALKAAQPVQVS
Mb1783c|M.bovis_AF2122/97 LVPRANPGP---------------NPAPNPPATG-PQPPNATQLTPNPAP
Rv1754c|M.tuberculosis_H37Rv LVPRANPGP---------------NPAPNPPATG-PQPPNATQLTPNPAP
MMAR_2586|M.marinum_M AVP-----------------------APAPPNPA-NQAPNATQIAPDAAP
MUL_3174|M.ulcerans_Agy99 AVP-----------------------APAPPNPA-NQAPNATQIAPDAAP
MLBr_02491|M.leprae_Br4923 --------------------------------------------------
MAV_1035|M.avium_104 EGPGPDPSAPMYPQA------EVLPPGPNPPTPGGAVPPNANQLRPNAAP
MAB_3454|M.abscessus_ATCC_1997 ELARP--------------------ALPNPPEPG---SPNNLQDQQAAAV
Mvan_5180|M.vanbaalenii_PYR-1 APQPPFLWALLSFARRELDNLFAARSTTNAPRTTSVTPTSLALTDDATTA
Mflv_1576|M.gilvum_PYR-GCK APQPPFLWALMAFARRELDTLFAAR-TSNSPRSSTAASTSLTLGDAATAA
TH_1246|M.thermoresistible__bu --------------------------------------------------
MSMEG_4365|M.smegmatis_MC2_155 PPRPTITGLVRGLLTAAAGFSPRAATAPVAPAPSPAAWALLAFARREFER
Mb1783c|M.bovis_AF2122/97 APDPAPAAAPD---PGATLAGATTSL-----------AEWVTGPD-----
Rv1754c|M.tuberculosis_H37Rv APDPAPAAAPD---PGATLAGATTSL-----------AEWVTGPD-----
MMAR_2586|M.marinum_M IPAPAPPPPPDGSDAGGALGGATTSL-----------AEWVTGPD-----
MUL_3174|M.ulcerans_Agy99 IPAPAPPPPPDGSDAGGALGGATTSL-----------AEWVTGPD-----
MLBr_02491|M.leprae_Br4923 ----------------------------------------MTGPN-----
MAV_1035|M.avium_104 MPQPQPQPQPAPGPPPPSIAGAPTSL-----------VDWVTGPN-----
MAB_3454|M.abscessus_ATCC_1997 APAP---PAPAVGPAADAAVSPTTSV-----------VGWVTGVD-----
Mvan_5180|M.vanbaalenii_PYR-1 ALIPS------YSPWLNPQVSPSTRF-----------VSWVTGKYVYLDD
Mflv_1576|M.gilvum_PYR-GCK ATAAATTPQPFRSPWLNPQVSASTKF-----------ISWVTGAFYDGDY
TH_1246|M.thermoresistible__bu -------------------------------------VDWITGPN-----
MSMEG_4365|M.smegmatis_MC2_155 AFTPGAATALAAPAITTSAVTSAAVVNPLPGRLDSVPVGWVTGVSNPSGN
:**
Mb1783c|M.bovis_AF2122/97 -SPNKTLERFGISGTDLGIPWDNG----DPAN-----RQVLMIFGDTFGY
Rv1754c|M.tuberculosis_H37Rv -SPNKTLERFGISGTDLGIPWDNG----DPAN-----RQVLMIFGDTFGY
MMAR_2586|M.marinum_M -SPNKTLERFGISGTDLGILWDNG----DPAN-----HQVLMAFGDTFGY
MUL_3174|M.ulcerans_Agy99 -SPNKTLERFGISGTDLGILWDNG----DPAN-----HQVLMAFGDTFGY
MLBr_02491|M.leprae_Br4923 -SPNKTLQRFGISGTDLGIPWDNG----DPTN-----HQVLMAFGDSFGY
MAV_1035|M.avium_104 -GPNKTLQRFSISGTDLGIAWDNG----DPAN-----RQVLMAFGDTFGY
MAB_3454|M.abscessus_ATCC_1997 -TGANTLQKFSISGTDLGIMWDNG----DNAG-----RQVLMAFGDTYGY
Mvan_5180|M.vanbaalenii_PYR-1 DTLANTLARFSVYGTDVGVMWDNGMVD-DPSTPDINEHQILIAVGDTFGA
Mflv_1576|M.gilvum_PYR-GCK ALLANTLKRFNVYGTDVGVMWDNGMVD-DPSTPDIDERQVLIAVGDTFGG
TH_1246|M.thermoresistible__bu ---STSYTRFGISGTDLGIMWDNGSAT-EP--------QVLIAFGDTFGD
MSMEG_4365|M.smegmatis_MC2_155 WPQTNNTAGFDIWGTDLGIMWDGGEWNG--------QRFVHTAFGDTFSG
.. *.: ***:*: **.* : .**::.
Mb1783c|M.bovis_AF2122/97 CAVDGHQWRYNTLFRSQDRDLGNGVHVTSGD--ASNRYSGSPVRQ--PGF
Rv1754c|M.tuberculosis_H37Rv CAVDGHQWRYNTLFRSQDRDLGNGVHVTSGD--ASNRYSGSPVRQ--PGF
MMAR_2586|M.marinum_M CAVREHQWRYNTLFRSQDRNLADGLHVTPGD--PVNRYSGSPISQ--PGF
MUL_3174|M.ulcerans_Agy99 CAVREHQWRYNTLFRSQDRNLADGLHVTPGD--PVNRYSGSPISQ--PGF
MLBr_02491|M.leprae_Br4923 CSVKGQQRRYNILFRSSNQDLSHGIRIADGV--PNDKYSGSPVWT--TGL
MAV_1035|M.avium_104 CKVHGQQWRYNTLFRSNDHDLSHGIHIADGV--PNDRYSGSPVWA--PGL
MAB_3454|M.abscessus_ATCC_1997 CGMKSQQWRYNTLLRTQDKSLSRGLAVAEGS--TTNPYAGSPQSR--PGY
Mvan_5180|M.vanbaalenii_PYR-1 KGMTGR-HIYNTLFRSSDTDLSDGMTIPDGEWFNGNMFGGAPLDG--PTQ
Mflv_1576|M.gilvum_PYR-GCK PGMTGR-HIYNSLFRSSDRDLADGMTIPDGEWFNGNMFGGAPLDK--PDQ
TH_1246|M.thermoresistible__bu CGVPGQEWRKNALFRSSDRNLADGMTIPDP--RYGDVYAGSPVTRERPDF
MSMEG_4365|M.smegmatis_MC2_155 PNMSGW-WRSNVLLISTDNVLSDGLGLVQTG----TAYQFIPASG-----
: * *: : : *. *: : : *
Mb1783c|M.bovis_AF2122/97 SKQLINSIKWARDETGIIPTAGIAV----------GKTQYVNFMSIRNWG
Rv1754c|M.tuberculosis_H37Rv SKQLINSIKWARDETGIIPTAGIAV----------GKTQYVNFMSIRNWG
MMAR_2586|M.marinum_M SKQIISSIKWAPDETGIIPTSGIAV----------GKTQYVSFMSIRDWG
MUL_3174|M.ulcerans_Agy99 SKQIISSIKWAPDETGIIPTSGIAV----------GKTQYVSFMSIRDWG
MLBr_02491|M.leprae_Br4923 AKQVVNTIHRAPHETGIIPTTAISI----------GKTQYMNYMSIKKWG
MAV_1035|M.avium_104 SKQVVNTIHKAAHETGIIPTSGMSM----------GRTQYMSYMSIRQWG
MAB_3454|M.abscessus_ATCC_1997 SKQIIPPIKWAAQERGIIPTAAISV----------GRTQYMNYMSIKSWD
Mvan_5180|M.vanbaalenii_PYR-1 ARPIINRPSWLPNSVTLIPTAGVSLPTEVTEDTPFGTIQYVSFMSVSNWG
Mflv_1576|M.gilvum_PYR-GCK SRPIIERPAWLPGSVTLIPTAGVSLPTEVSEDTPFGTIQYVSFMSVTRWG
TH_1246|M.thermoresistible__bu SRQVIASLGLAAQEVTVIPTAGISV----------GSTQYVNFMSVAQWG
MSMEG_4365|M.smegmatis_MC2_155 -RNLFG--LFGGSEVTVIPTAGVQIDG----------TQYVNYMSVRSWD
: :. . :***:.: : **:.:**: *.
Mb1783c|M.bovis_AF2122/97 RDGEWTTNYSGIAVSKDNGQTWGVFPGTIRASGPDSGGKARFVPGNENFQ
Rv1754c|M.tuberculosis_H37Rv RDGEWTTNYSGIAVSKDNGQTWGVFPGTIRASGPDSGGKARFVPGNENFQ
MMAR_2586|M.marinum_M RDGEWTTNYSGIAVSKDNGQNWGVFPGTIRASGPDSG-RVRFVPGNENFQ
MUL_3174|M.ulcerans_Agy99 RDGEWTTNYSGIAVSKDNGQNWGVFPGTIRASGPDSG-RVRFVPGNENFQ
MLBr_02491|M.leprae_Br4923 RDGEWTTNYSAIARSIDNGQSWGTYPGTIRTASPDAIPGTHFVPGNQNFQ
MAV_1035|M.avium_104 RDGEWSTNYSAIARSTDNGQNWGIFPTSIRTASPDAIPGAGFTPGNENFQ
MAB_3454|M.abscessus_ATCC_1997 SAGEWTTNYSATAVSNDNGQNWKTFPQSIRPASPDAISQVPFTPGNENFQ
Mvan_5180|M.vanbaalenii_PYR-1 SSGRWTTNYSAIAYSTDNGENFKVAPESVR---YN-----SIFSGNRNFQ
Mflv_1576|M.gilvum_PYR-GCK QSGRWSTNYSGIAYSTDNGENFTVAPSSVR---YN-----SFFSGNRNFQ
TH_1246|M.thermoresistible__bu APGQWSTNFSAVAVSTDNGETWTVPRESIRPAWFNSVPGVPFIWGYQNFQ
MSMEG_4365|M.smegmatis_MC2_155 TPGRWTTNFSAISVYDPSTDTWVLAPSTIRSAGWFRS-STLYVPGSQNFQ
*.*:**:*. : . :.: ::* * .***
Mb1783c|M.bovis_AF2122/97 MGAYLKSNDG--------YLYSFGTPPGRGGSAYLARVPQRFVPDLTKYQ
Rv1754c|M.tuberculosis_H37Rv MGAYLKSNDG--------YLYSFGTPPGRGGSAYLARVPQRFVPDLTKYQ
MMAR_2586|M.marinum_M MGAFVKANDG--------YLYSFGTPPGRGGSAYLARVPQRFVPDLTKYQ
MUL_3174|M.ulcerans_Agy99 MGAFVKANDG--------YLYSFGTPPGRGGSAYLARVPQRFVPDLTKYQ
MLBr_02491|M.leprae_Br4923 MGAFMRGNDG--------YLYSFGTPSGRSGAAYLARVPQNLVPDLSKYQ
MAV_1035|M.avium_104 MGAFLKGNDG--------YIYQFGTPSGRGGAAFLSRVPPNQLPDLTKYQ
MAB_3454|M.abscessus_ATCC_1997 QAAYVKGNDG--------YIYIFGTPSGRSGSGFVSRALPGNLPDAAKHE
Mvan_5180|M.vanbaalenii_PYR-1 QSAFVKGDDG--------YVYVYGTPNGRQGAAYVARVNNENILDVSEYE
Mflv_1576|M.gilvum_PYR-GCK QSAFVKGDDG--------YVYMYGTPNGRQGAAYLARVAPEDILNASKYE
TH_1246|M.thermoresistible__bu MGAYLR-HNG--------YVYAYGTGAGRGGMPFLARVPEHSVANLAAYE
MSMEG_4365|M.smegmatis_MC2_155 QAAYVLQPEDQVGEDGIRYLYAFGTPSGRAGSAYLSRVPEDAVTDLSKYE
.*:: :. *:* :** ** * :::*. : : : ::
Mb1783c|M.bovis_AF2122/97 YWNGDSNS--------WVPNKPDAATPVIPGPVG----------------
Rv1754c|M.tuberculosis_H37Rv YWNGDSNS--------WVPNKPDAATPVIPGPVG----------------
MMAR_2586|M.marinum_M YWNGDSNS--------WVAGKPGAATPVIPGPVG----------------
MUL_3174|M.ulcerans_Agy99 YWNGDSNS--------WVAGKPGAATPVIPGPVG----------------
MLBr_02491|M.leprae_Br4923 YWNGN-----------WVPNNPGAATPLFSGPVG----------------
MAV_1035|M.avium_104 YWNGDNGGH-------WVANNPGAATPIFPGPVG----------------
MAB_3454|M.abscessus_ATCC_1997 FWNTDRGT--------WVPGDPNAATPILSGPVG----------------
Mvan_5180|M.vanbaalenii_PYR-1 YYKAASTGWFGSSEARWVKGNPSSASAVIGKTGGACGST-----------
Mflv_1576|M.gilvum_PYR-GCK YYKKASSGWFGGSPARWV-GSPSSASAIIGKSGGLFGFT-----------
TH_1246|M.thermoresistible__bu YHTPFG----------WLRGLPFLAIQVVWAPS-----------------
MSMEG_4365|M.smegmatis_MC2_155 YWDGNR----------WVTGNAAVAAPVIGDSTRSTGLFGFIVDWANDPN
: *: . . * :. .
Mb1783c|M.bovis_AF2122/97 -------------------EMSVQYNTYLKQYLALYTNG-MNDVVARTAP
Rv1754c|M.tuberculosis_H37Rv -------------------EMSVQYNTYLKQYLALYTNG-MNDVVARTAP
MMAR_2586|M.marinum_M -------------------EMSAQYNTYLKQYLVMYTNG-GNDVVARTAP
MUL_3174|M.ulcerans_Agy99 -------------------EMSAQYNTYLKQYLVMYTNG-GNDVVARTAP
MLBr_02491|M.leprae_Br4923 -------------------EMSAQYNDYLKQYIILYSNGDSNDVVARTAP
MAV_1035|M.avium_104 -------------------EMSAQYNSYLKQYLVLYTDGGSNDVVARTAP
MAB_3454|M.abscessus_ATCC_1997 -------------------EMSAQYNTYLKKYLVMYGDK-SGDVLLSTSP
Mvan_5180|M.vanbaalenii_PYR-1 ------------EPGYTVSEMSVQYNEYLQKYIVLYGDQ-FNNIVMRTSD
Mflv_1576|M.gilvum_PYR-GCK ------------KPGYTVSEMSVQYNEYLKKYIVLYGDQ-SNNIVMRTSD
TH_1246|M.thermoresistible__bu ------------------SELSVAWNDYLQKFVMLYTNT-ASSVVMRTAD
MSMEG_4365|M.smegmatis_MC2_155 VLGGYLGGLVGAKTGGNVSELSVQYNEYLGKYVVLYGDG-NNNIQMRIAD
*:*. :* ** ::: :* : ..: :
Mb1783c|M.bovis_AF2122/97 APQGPWSAEQMLVSSWQMPGGIYAPMMHPWS------------TGKDVYF
Rv1754c|M.tuberculosis_H37Rv APQGPWSAEQMLVSSWQMPGGIYAPMMHPWS------------TGKDVYF
MMAR_2586|M.marinum_M TPQGPWSPEQMLVSSWQMPGGIYAPMMHPWS------------TGKDVYF
MUL_3174|M.ulcerans_Agy99 TPQGPWSPEQMLVSSWQMPGGIYAPMMHPWS------------TGKDVYF
MLBr_02491|M.leprae_Br4923 APQGPWSPEQPLVSSFQMPGGIYAPMIHPWS------------SGRDLYF
MAV_1035|M.avium_104 TPQGPWSPEQPLVSSFQMPGGIYAPMIHPWS------------SGRDLYF
MAB_3454|M.abscessus_ATCC_1997 APQGPWSPPQIIVTESQMPGGPYAPYLHPWT------------TGKELYF
Mvan_5180|M.vanbaalenii_PYR-1 TPEGAWSDATVLMG--QQSGGIYAPMLHPWSPS-------TQGTGSDLYW
Mflv_1576|M.gilvum_PYR-GCK TPEGGWSDATVLMR--QQTGGIYAPMLHPWSPS-------TEGTGSTLYW
TH_1246|M.thermoresistible__bu KPEGPWSAAKTIVTSTQVPGGIYAPYIHPWS------------KGRDLYF
MSMEG_4365|M.smegmatis_MC2_155 RPEGPWSDPIELASSADYPG-LYAPMIHPWSGTGMLEDDNGDPDVNNLYW
*:* ** : : .* *** :***: :*:
Mb1783c|M.bovis_AF2122/97 NLSLWSAYNVMLMHTVLP-------
Rv1754c|M.tuberculosis_H37Rv NLSLWSAYNVMLMHTVLP-------
MMAR_2586|M.marinum_M NLSLWSAYNVMLMHTELA-------
MUL_3174|M.ulcerans_Agy99 NLSLWSAYNVMLMHTELA-------
MLBr_02491|M.leprae_Br4923 NLSLWSAYDVVLMHTVLP-------
MAV_1035|M.avium_104 NLSLWSAYDVMLMHTVLP-------
MAB_3454|M.abscessus_ATCC_1997 NLSLWSAYNVMLMRTTLP-------
Mvan_5180|M.vanbaalenii_PYR-1 NLSLWSDYNVMLMKTDLTKL-----
Mflv_1576|M.gilvum_PYR-GCK NLSLWSEYNIMLMETDLSKI-----
TH_1246|M.thermoresistible__bu TLSLWSTYSVMLMRTTLQK------
MSMEG_4365|M.smegmatis_MC2_155 NMSLWGDYNVVLMQTDLSGLKTVAV
.:***. *.::**.* *