For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALAGMSLGTDLKRYSRGLMPRIALITIILMPLLYGAMYLWAFWNPFAEVNKVPVALVNEDRGASAQGQPL RAGDDVATALMQSGQLDLTEVSAQQAADGVARGDYYFSITLPPDFSERVASPSGDDPQQAQIRFTFNDAN NYLATIIGQNASREVLNQVNAKIGERTVGTVLTGLTDAGQGLEQAADGAQQLATGLDTADDGAQRLATGA GTLSSGLDSARAGSAQLAAGTRQLAGTVDTATGPLLEMLDRVAGLSLNPDEVAVAAQHLSGAVRSTTDRI AALNVNSAQAAAIVDQAVGFLQANPDPAVRDAGHALAGAQRLLRAQNIDPTTDEGLIRLRDSASRLEGEL GDPNSKLRVFLARALDGGLRSDVAKLRDGVDELNSGAQRLNAGLVQLANGGRELSSGASQLADGTEKLRA GGQELATKLRQGTAEVPSWTPQQRTEMAQTLVTPVGLDLVNTNPAATFGTGFAPFFMPLALFIGALIIWM LLTPLQSRPIVNGLGALRVVLASYWPGLLVAVCQVVVMYAVVHFGVGLQAEYPLATVAFLVLIAATFLAM IQAFNAVFGVAVGRVVTLAFLMLQLVSAGGIYPVETTAKPFQIIHPFDPMTYAVNGLRQLTVGGVDSRLW IAIAVLAGLLVASLAASAWSARRNRQYTMERLHPPIEV*
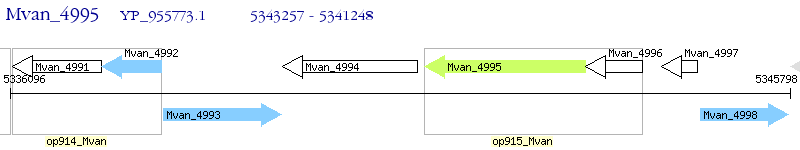
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4995 | - | - | 100% (669) | ABC-2 type transporter |
| M. vanbaalenii PYR-1 | Mvan_0312 | - | 4e-11 | 23.45% (516) | MMPL domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_2900 | - | 2e-10 | 24.47% (523) | MMPL domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0186c | - | 3e-10 | 27.98% (243) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_1752 | - | 0.0 | 82.81% (669) | ABC-2 type transporter |
| M. tuberculosis H37Rv | Rv0180c | - | 4e-10 | 27.98% (243) | transmembrane protein |
| M. leprae Br4923 | MLBr_02600 | - | 3e-07 | 26.34% (243) | hypothetical protein MLBr_02600 |
| M. abscessus ATCC 19977 | MAB_4762 | - | 1e-156 | 43.71% (668) | hypothetical protein MAB_4762 |
| M. marinum M | MMAR_0423 | - | 5e-09 | 25.93% (243) | transmembrane protein |
| M. avium 104 | MAV_1613 | - | 1e-155 | 44.68% (667) | ABC-2 type transporter family protein |
| M. smegmatis MC2 155 | MSMEG_5644 | - | 0.0 | 76.31% (667) | membrane protein |
| M. thermoresistible (build 8) | TH_0090 | - | 0.0 | 74.90% (514) | membrane protein |
| M. ulcerans Agy99 | MUL_1073 | - | 2e-09 | 27.05% (244) | transmembrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4995|M.vanbaalenii_PYR-1 MALAGMSLGTDLKRYSRGLMPRIALITIILMPLLYGAMYLWAFWNPFAEV
Mflv_1752|M.gilvum_PYR-GCK MALAGMSLGTDLSRYFHGRLPRIALATIIVMPLLYGAMYLWAFWNPFAEV
MSMEG_5644|M.smegmatis_MC2_155 -MLAGMSLGTDFKRFSRGAMPRIALVIVIFMPLLYGAMYLWAFWNPFNEV
TH_0090|M.thermoresistible__bu --------------------------------------------------
MAB_4762|M.abscessus_ATCC_1997 -MLAGLAFGSEIKRFGRSRMTRAAIVVLMLLPLVYGALYLWAYWDPFGHV
MAV_1613|M.avium_104 -MLAGLAFGSEIKRFRRSRLTRAAIVVLMLLPLVYGALYLWAFWDPFGHT
Mb0186c|M.bovis_AF2122/97 --------------------------------------------------
Rv0180c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0423|M.marinum_M --------------------------------------------------
MUL_1073|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02600|M.leprae_Br4923 --------------------------------------------------
Mvan_4995|M.vanbaalenii_PYR-1 NKVPVALVNEDRGASAQGQPLRAGDDVATALMQSGQLDLTEVSAQQAADG
Mflv_1752|M.gilvum_PYR-GCK NKVPVALVNEDRGASAQGQDLRAGDEVARALIDSGQLHLTEMSAQEAAEG
MSMEG_5644|M.smegmatis_MC2_155 DKVPVALVNEDKGAVAQGERLNAGQEVSDALVDSGELQLHRVSADEAADG
TH_0090|M.thermoresistible__bu --------------------------------------------------
MAB_4762|M.abscessus_ATCC_1997 NKMPVALVNADKGATVSGQHVNIGEEISKSLTADGSMDWHVLSLDEARSG
MAV_1613|M.avium_104 NKMPVALVNSDRGAVVSGQQFNAGAEIAKSLTADGSLDWHVVDLPEARNG
Mb0186c|M.bovis_AF2122/97 --------------------------------------------------
Rv0180c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0423|M.marinum_M --------------------------------------------------
MUL_1073|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02600|M.leprae_Br4923 --------------------------------------------------
Mvan_4995|M.vanbaalenii_PYR-1 VARGDYYFSITLPPDFSERVASPSGDDPQQAQIRFTFNDANNYLATIIGQ
Mflv_1752|M.gilvum_PYR-GCK VAAGDYYFSITLPEDFSASVASPSGGDPRQATIRFTFNDANNYLASVIGQ
MSMEG_5644|M.smegmatis_MC2_155 VSSGRYYFSITLPEDFSEAIASPSGDSPQQATLRFTFNDANNYLGSIIGQ
TH_0090|M.thermoresistible__bu --------------------------------------------------
MAB_4762|M.abscessus_ATCC_1997 VDHGNYYFMLELPPDFSEAIASPLTGEPKQANLVAVYNDANNYISSSIGR
MAV_1613|M.avium_104 VDHGKYYFMVELPPDFSAAIASPVTGQPKKANLIAVYNDANNYISTSIGR
Mb0186c|M.bovis_AF2122/97 --------------------------------------------------
Rv0180c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0423|M.marinum_M --------------------------------------------------
MUL_1073|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02600|M.leprae_Br4923 --------------------------------------------------
Mvan_4995|M.vanbaalenii_PYR-1 NASREVLNQVNAKIGERTVGTVLTGLTDAGQGLEQAADGAQQLATGLDTA
Mflv_1752|M.gilvum_PYR-GCK NASREIVNQVNAEIGERSVGTVVTGLTDAGQGLQQAADGAQQLATGLGTA
MSMEG_5644|M.smegmatis_MC2_155 NAAREVINQINARIGERTVGTVLTGLTDAGEGLIKAADGAQQLADGNAAA
TH_0090|M.thermoresistible__bu -----VLNVVNARVGQQSVETVLTGLTDAGEGLTQAADGADRLHTGLVAA
MAB_4762|M.abscessus_ATCC_1997 TAIDQVLNAVSTRISGQAVNQVLSVVVSSGAGIQQAADGARQLADGAAKV
MAV_1613|M.avium_104 TAIEQVLNAVSSRISGQAVNQMLSVVVSSGSGIKQAAD-------GAARL
Mb0186c|M.bovis_AF2122/97 ------MSQAQPRPAAPNPKRNVKAIR-----------------------
Rv0180c|M.tuberculosis_H37Rv ------MSQAQPRPAAPNPKRNVKAIR-----------------------
MMAR_0423|M.marinum_M ------MSQSQPRHAAPNPKRSIKSLR-----------------------
MUL_1073|M.ulcerans_Agy99 ------MSQSQPRHAAPNPKRSIKSLR-----------------------
MLBr_02600|M.leprae_Br4923 ------MSQPQPRHAPFAPTRDAKAIR-----------------------
:. ... . . :
Mvan_4995|M.vanbaalenii_PYR-1 DDGAQRLATGAGTLSSGLDSARAGSAQLAAGTRQLAGTVDTATGPLLEML
Mflv_1752|M.gilvum_PYR-GCK DDGARRLAGGADTLSSGLNTAQDGSAELAAGTQRLATSIDKTTGPLLEML
MSMEG_5644|M.smegmatis_MC2_155 NDGAQRLASGADALTAGLVTARDGSAQLAAGTRQLSTAVTTATDPLLHVL
TH_0090|M.thermoresistible__bu DDGAHDLSVGADALAAGLVTARDGAAQLAAGTGQLAAAVDTAIDPLLHTL
MAB_4762|M.abscessus_ATCC_1997 DD-------GAGQLATGLHTARPGSAQLATGAKQLSDGINQATDPLLTVT
MAV_1613|M.avium_104 DD-------GAGQLAAGLDSARTGSATLAAGAKQLSDGINQATDPLLAVT
Mb0186c|M.bovis_AF2122/97 ------------TVRFWMAPIATTLALMSALAALYLGGILNPMTNLRHFP
Rv0180c|M.tuberculosis_H37Rv ------------TVRFWMAPIATTLALMSALAALYLGGILNPMTNLRHFP
MMAR_0423|M.marinum_M ------------TVRFWTAPVVITLALMSALAALYLGGILNPTTNLRQFP
MUL_1073|M.ulcerans_Agy99 ------------TVRFWTAPVVIILALMSALAALYLGGILNPTTNLRQFP
MLBr_02600|M.leprae_Br4923 ------------TVRFWASPIVITLSLMSALAALYLGGILNPTTNLQHFP
: . : ::: : : . *
Mvan_4995|M.vanbaalenii_PYR-1 DRVAGLSLNPDEVAVAAQHLSGAVRSTTDRIAALNVNSAQAAAIVDQAVG
Mflv_1752|M.gilvum_PYR-GCK DRVGAVGLNPDDVAVAAQHLSGAVRSTTDRLAALNVNTAQAAAIVDHSVG
MSMEG_5644|M.smegmatis_MC2_155 DRVGDLGLDPDEVGRAAQRLSGAVRSTTDRIAALNIDQAQAAAIVDQAVN
TH_0090|M.thermoresistible__bu DRVGALGLDPDEVGAVATRLSGAVRSTTERIAALHIDQHHAAAVLDQAVT
MAB_4762|M.abscessus_ATCC_1997 KAVANIGGSTDKLQQGAEALRQAN----DQIDGIAKAQDSAANALTAVID
MAV_1613|M.avium_104 KAVSQIGGSTEQLQQGATALAQAN----DQLGAIAAAQDAAASSLTSVID
Mb0186c|M.bovis_AF2122/97 IALVNEDAGPAGQQIVDGLVSGLDK---NKFDIRVVSPDEARRLLDTAAV
Rv0180c|M.tuberculosis_H37Rv IALVNEDAGPAGQQIVDGLVSGLDK---NKFDIRVVSPDEARRLLDTAAV
MMAR_0423|M.marinum_M IALVNEDAGPAGKQIVDGIVAGLDK---NKFDVRVVDHEQAQRLLDRAQV
MUL_1073|M.ulcerans_Agy99 IALVNEDAGPAGKQIVDGIVAGLDK---NKFDVRVVDHEQAQRLLDRAQV
MLBr_02600|M.leprae_Br4923 LAVVNEDAGPAGAQIVEGLLAGLDK---NKFDVRVVSCREAQQLLDSAGV
: . .. : ::: * :
Mvan_4995|M.vanbaalenii_PYR-1 FLQANPDPAVRDAGHALAGAQRLLRAQNIDPTTDEGLIRLRDSASRLEGE
Mflv_1752|M.gilvum_PYR-GCK FLQSSPDPAVRDAGHALAGAQRLLRAQGIDPTTDEGLIRLRDSASRLESE
MSMEG_5644|M.smegmatis_MC2_155 FLRNSPDPTARDTGEFLAGAHRILTSRGIDPTTDDGLIKLRDSAAELENE
TH_0090|M.thermoresistible__bu FLRTNPDPAAHHLGEALAGVQRMLRAQGIDPATDEGLIRLRDSAAQLESE
MAB_4762|M.abscessus_ATCC_1997 QLAGRQDPAAN----TLRGIQDQLREHQFTPQVRQQLTDAENASIAMTET
MAV_1613|M.avium_104 QLSARADPLAN----NLRGIQDQLRGHQFTPQIRQQLTDAQNAAIAMTSG
Mb0186c|M.bovis_AF2122/97 YGSALIPPTFS------SQLRDFGASAVTPTRTDRPAITISTNPRAGTLA
Rv0180c|M.tuberculosis_H37Rv YGSALIPPTFS------SQLRDFGASAVTPTRTDRPAITISTNPRAGTLA
MMAR_0423|M.marinum_M YGSAVIPPTFS------SQLRDFAVSAVQPGSTDRPVITISTNPRAGTLG
MUL_1073|M.ulcerans_Agy99 YGSAVVPPTFS------SQLRDFAVSAVQPGSTDRPVITIFTNSRAGTLG
MLBr_02600|M.leprae_Br4923 YGSAVIPPSFS------SQLRDFGTSAVLPTHADRPVIMVSTNPRAGALG
* : .
Mvan_4995|M.vanbaalenii_PYR-1 LGDPNSKLRVFLARALDGGLRSDVAKLRDGVDELNSGAQRLNAGLVQLAN
Mflv_1752|M.gilvum_PYR-GCK LGDPQSTLRVFLARALNGGLRSDVASLRDGVDQLNSGAQRLADGLVQLAG
MSMEG_5644|M.smegmatis_MC2_155 LGDPNSKMRTFISKALNGGLRADVVKLRDGAQQLNSGAQALNSGLVQLTN
TH_0090|M.thermoresistible__bu LADPNSRLRTFLTAALDGGLRGEVVRLRDGVNRLDTGARQLSAGLVRLAD
MAB_4762|M.abscessus_ATCC_1997 LRGPGSPLKSALDQVGGKGQ-----ELTNKLTQLRNGAQQLATGNAQLSS
MAV_1613|M.avium_104 LRTPGSPLASALDQLGGKGQ-----ELTNKLTQLRDGAQQVATGNAELAG
Mb0186c|M.bovis_AF2122/97 ASIAGQTLTRALTVVNGKVG----ERLTAEVAAQTGGVALAGAAAAGLAS
Rv0180c|M.tuberculosis_H37Rv ASIAGQTLTRALTVVNGKVG----ERLTAEVAAQTGGVALAGAAAAGLAS
MMAR_0423|M.marinum_M ASIAGQTLTQAMAAANSKVG----ERVTAEVAAQTGGAPLSGASALGLAS
MUL_1073|M.ulcerans_Agy99 ASIAGQTLTQAMAAANSKVG----ERVTAEVAAQTGGAPLSGASALGLAS
MLBr_02600|M.leprae_Br4923 SSIAGQTLTQAITMVNSKAG----QRLTAEVAAQTGGAHLTGAAALGLTS
. . : : . : *. . *:.
Mvan_4995|M.vanbaalenii_PYR-1 GGRELSSGASQLADGTEKLRAGGQELATKLRQGTAEVPSWTPQQRTEMAQ
Mflv_1752|M.gilvum_PYR-GCK GGRELAAGAHDLAEGTGKLRAGGQELATRLRDGSTQIPSWTPQQRTEVAR
MSMEG_5644|M.smegmatis_MC2_155 GGRQLSNGANELAAGTEKLETGSRQLATALRDGSTQVPSWTPQQRTEVAR
TH_0090|M.thermoresistible__bu GGQQLATGAGRLADGTGQLRAGSRELADALRDGSTQVPSWTPEQRSDVAA
MAB_4762|M.abscessus_ATCC_1997 GIAKMDDGAQQLKSGTAQLRSGSAELATKLTDGAKQVPTWSNQQKNAIAD
MAV_1613|M.avium_104 GIAKLDDGAGQLKAG-------SAELATKLAEGARQVPNWTTQQKEAVAD
Mb0186c|M.bovis_AF2122/97 PIDVKSTAYNPLPNGTG------NGLSAFYYALLLLLAGFTG--SIVVST
Rv0180c|M.tuberculosis_H37Rv PIDVKSTAYNPLPNGTG------NGLSAFYYALLLLLAGFTG--SIVVST
MMAR_0423|M.marinum_M PIDVKTTVYNPLPNGTG------NGLSAFYYALLLLLAGFTG--SIVVST
MUL_1073|M.ulcerans_Agy99 PIDVKTTVYNPLPNGTG------NGLSAFYYALLLLLAGFTG--SIVVST
MLBr_02600|M.leprae_Br4923 PIDIKLAIYNPLPNGTG------NGLSAFYYALLLLLTGFTG--SIVVST
* * *: :. :: ::
Mvan_4995|M.vanbaalenii_PYR-1 TLVTPVGLDLVNTNPAATFGTGFAP-FFMPLALFIGALIIWMLLTPLQSR
Mflv_1752|M.gilvum_PYR-GCK TLAAPVGADLVTDNPAPTFGTGFAP-FFLPLALFIGALIVWMLLSPLRSR
MSMEG_5644|M.smegmatis_MC2_155 TLSAPVALDIETHNPAVTFGTGFAP-FFLPLALFIGALIIWMLLKPLQTR
TH_0090|M.thermoresistible__bu AMAAPVELDLVTHNPAATFGTGFAP-FFLPLALFIGALIVWMLLTPLQSR
MAB_4762|M.abscessus_ATCC_1997 TIGGPVHLETAHENAAPNFGTGMAP-FFVTLALFFGALVLWMILRPLQTR
MAV_1613|M.avium_104 TIGGPVQLEASHENAAPNFGTGMAP-FFLTLALFFGALVLWMVLRPLQYR
Mb0186c|M.bovis_AF2122/97 LVDSMLGYVPAEFGPVYRFAEQVNISRFRTLLVKWAVMVVLALLTSGVYL
Rv0180c|M.tuberculosis_H37Rv LVDSMLGYVPAEFGPVYRFAEQVNISRFRTLLVKWAVMVVLALLTSGVYL
MMAR_0423|M.marinum_M LVDSLLGYVPAEFGPVYRFAEQVNISRFRTLLLKWGLMTVLALLTSAVYM
MUL_1073|M.ulcerans_Agy99 LVDSLLGYVPAEFGPVYHFAEQVNISRFRTLLLKWGLMTVLALLTSAVYM
MLBr_02600|M.leprae_Br4923 LVDSMLGYIPTEVGPVYHYAEQINISRFHTLVVKWGLMVVLALITSAVYL
: : ... :. . * .* : . : : :: .
Mvan_4995|M.vanbaalenii_PYR-1 PIVNGLGALRVVLASYWPGLLVAVCQVVVMYAVVHFGVGLQAEYPLATVA
Mflv_1752|M.gilvum_PYR-GCK PIVNGLGALRVVLASFWPALLVVVCQVFVMYVVVHFGVGLQAKHPVATVG
MSMEG_5644|M.smegmatis_MC2_155 PVVNGLGALRVVLASYWPGLLIAVCQVVVMYVVVHFGVGLQAKYPLATVG
TH_0090|M.thermoresistible__bu PIVNGLGALRVVLASFWPGLLIVVAQVVVMYAVVHFGVGLKAEHPLATVA
MAB_4762|M.abscessus_ATCC_1997 AIAAEVLPLRVALSSYLPAATIGIFQAIILYCVVRFALGMHAAHPVAMLG
MAV_1613|M.avium_104 AIAAEVIAIRVVLASYLPAAAIALFQAVVLYCVVRFALGMHAVHPVAMLG
Mb0186c|M.bovis_AF2122/97 AIAHGLGMPIPLGWQVWLYGVFAIIAVGVTSSSLIAVLGSMG--LLVSML
Rv0180c|M.tuberculosis_H37Rv AIAHGLGMPIPLGWQVWLYGVFAIIAVGVTSSSLIAVLGSMG--LLVSML
MMAR_0423|M.marinum_M AIAHGLGMPIPAGWKLWLYGVFAITAVGVTSSSLIAVLGSMG--LLASML
MUL_1073|M.ulcerans_Agy99 AIAHELGMPIPAGWKLWLYGVFAITAVGVTSSSLIAVLGSMG--LLASML
MLBr_02600|M.leprae_Br4923 AIAHGLGMPIPLGWQLWLYGVFAISAVGITSSSLIAGLGSMG--LLVSMF
.:. : . . : . : : :* . :. :
Mvan_4995|M.vanbaalenii_PYR-1 FLVLIAATFLAMIQAFNAVFGVAVGRVVTLAFLMLQLVSAGGIYPVETTA
Mflv_1752|M.gilvum_PYR-GCK FLVLIGGTFLALIQAFNALFGVAVGRVVTLAFLMLQLVSAGGIYPVETTA
MSMEG_5644|M.smegmatis_MC2_155 FLILVAATFLALIQAFNALFGVSVGRVVTLAFLMLQLVSAGGIYPVETTA
TH_0090|M.thermoresistible__bu FLILIGATFLALIQAFNAVFGVAVGRVVTLAFLMLQLVSAGGIYPVETTA
MAB_4762|M.abscessus_ATCC_1997 FMVLISFAFVAATQAINALVGPAVGRVLLMALLMLQLVSAGGMYPVETTS
MAV_1613|M.avium_104 FMVLISAAFVAATQAINALVGPAVGRVLIMALLMLQLVSAGGMYPVETTS
Mb0186c|M.bovis_AF2122/97 IFVILGLPSAGATVPLEAVPAFFRWLAQFEPMHQVFLGVRSLLYLNGNAD
Rv0180c|M.tuberculosis_H37Rv IFVILGLPSAGATVPLEAVPAFFRWLAQFEPMHQVFLGVRSLLYLNGNAD
MMAR_0423|M.marinum_M VFVILGLPSAGATVPLEAVPPFFRWLAKFEPMHQVFLGVRSLLYLNGQPE
MUL_1073|M.ulcerans_Agy99 VFVTLGLPSAGATVPLEAVPPFFRWLAKFEPMHQVFLGVRSLLYLNGQPE
MLBr_02600|M.leprae_Br4923 IFMILGLPSAGATVPLEAVPPFFRWLSEFEPLHQLFLGVRSLLYLKGHAD
.:: :. . . .::*: .: : * . :* .
Mvan_4995|M.vanbaalenii_PYR-1 KPFQIIHPFDPMTYAVNGLRQLTVGGVDSRLWIAIAVLAGLLVASLAASA
Mflv_1752|M.gilvum_PYR-GCK KPFQILHPFDPMTYAVNGLRQLTVGGVDSRLWVGIGVLAGLLAASLATSA
MSMEG_5644|M.smegmatis_MC2_155 KPFQILHPVDPMTYAVNGLRQVIVGGVDSRLWIAIVVLTGLLAVSLAGSA
TH_0090|M.thermoresistible__bu KPFQIIHPFDPMTYAVNGLRQLIVGGVDQRLWIAIAVLAGLLAASTAASA
MAB_4762|M.abscessus_ATCC_1997 RPFQILHKYDPMTYGVNGLRQLILGGIDGRLWQAVITLLFILLGGLLITS
MAV_1613|M.avium_104 RPFQVLHRFDPMTYGVNGLRQLILGGIDARLWQAIIVLAAITAVALAISC
Mb0186c|M.bovis_AF2122/97 AGLSQALTMTSIGLIIG----LLLGGFITHLYDRSSFHRIPGAVEMAIAV
Rv0180c|M.tuberculosis_H37Rv AGLSQALTMTSIGLIIG----LLLGGFITHLYDRSSFHRIPGAVEMAIAV
MMAR_0423|M.marinum_M SGLSHALLMTAIGLVIG----LLLGGLTTHLYDRKGFHRIPGAVEMAIAA
MUL_1073|M.ulcerans_Agy99 SGLSHALLMTAIGLVIG----LLLGGLTTHLYDRKGFHRIPGAVEMAIAA
MLBr_02600|M.leprae_Br4923 SGMSQALFMTALGMAIG----LLLGGVVTRIYDRGGCHRIPGAVGMAIAE
:. .: :. : :**. ::: :
Mvan_4995|M.vanbaalenii_PYR-1 WSARRNRQYTMER----------LHPPIEV---
Mflv_1752|M.gilvum_PYR-GCK WAARRNRQYTVDR----------LRPPIEV---
MSMEG_5644|M.smegmatis_MC2_155 WAARRNRQYTMDR----------LRPPIEV---
TH_0090|M.thermoresistible__bu WAARRNRQYTMQR----------LHPPIEV---
MAB_4762|M.abscessus_ATCC_1997 LSARRNQLWNLTR----------LLPSIKM---
MAV_1613|M.avium_104 LSARRDRLWNLSR----------LFPAIKI---
Mb0186c|M.bovis_AF2122/97 EHQAQYQARQSAR-----------ESSSEQP--
Rv0180c|M.tuberculosis_H37Rv EHQAQYQARQSAR-----------ESSSEQP--
MMAR_0423|M.marinum_M EHQAQHQAKERARDRD------STESSSEET--
MUL_1073|M.ulcerans_Agy99 EHQAQHHAKERARDRD------STESSSEET--
MLBr_02600|M.leprae_Br4923 WHQAQHQARLRPNKSGKRPNVIPTQPKCHLPVI
: : . . .