For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ANFSVPKGRLMLNGGAGSKAASHVADEPLTEAATSDVVLERRITLGRGPVGDIAAGQGFVVMANAGDDSV SVIDPMTLAVVDTIAVDGQPVAVTASDDRAYVCVAAESHDAVCAIDLEAGTVVSTHPLASGVTALAVSPD GKRVYAGRDAQDRVDVAVIDVTAERVGTIGIGSGPAANVDALSVDPSGKRLYVAVTDAVGSRLVIVDTET SLVHRVLPIGAPIRDIAYAGGAVYVLTSDRAVGGTVHVVDLNAMRVTDRVVVGGAPTQLTMSADQTRAYI VDYDRVAVLCTLGLDVVDSLAVDARPSCVTHPADSSRLYIADYTGAVSVFSVESSIETLYSQFLATDSIA LGVSRALQPTPA*
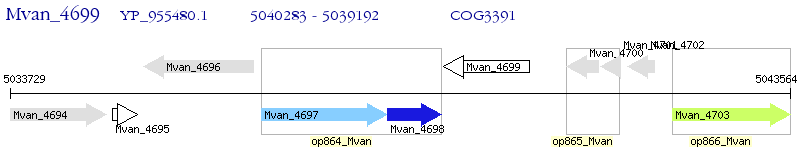
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4699 | - | - | 100% (363) | hypothetical protein Mvan_4699 |
| M. vanbaalenii PYR-1 | Mvan_3594 | - | 8e-15 | 25.86% (290) | YVTN beta-propeller repeat-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1720 | - | 3e-05 | 21.51% (344) | YVTN beta-propeller repeat-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1086 | - | 2e-42 | 30.92% (359) | hypothetical protein Mb1086 |
| M. gilvum PYR-GCK | Mflv_2024 | - | 1e-125 | 67.17% (329) | hypothetical protein Mflv_2024 |
| M. tuberculosis H37Rv | Rv1057 | - | 2e-42 | 30.92% (359) | hypothetical protein Rv1057 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4413 | - | 1e-41 | 32.48% (351) | hypothetical protein MMAR_4413 |
| M. avium 104 | MAV_1185 | - | 2e-40 | 33.87% (313) | hypothetical protein MAV_1185 |
| M. smegmatis MC2 155 | MSMEG_5308 | - | 3e-85 | 50.31% (320) | hypothetical protein MSMEG_5308 |
| M. thermoresistible (build 8) | TH_4450 | - | 1e-68 | 47.39% (306) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_0230 | - | 4e-41 | 32.48% (351) | hypothetical protein MUL_0230 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4699|M.vanbaalenii_PYR-1 --MANFSVPKGRLMLNGGAGSKAASHVADEPLTEA-----ATSDVVLERR
Mflv_2024|M.gilvum_PYR-GCK -------------MHNVHNRSSAEDTVR--------------GDIVRERV
MSMEG_5308|M.smegmatis_MC2_155 MTMAKNILRAITAIVRRDAKTPDTDDSAGSVTFDG-----ALDDLDVAGL
TH_4450|M.thermoresistible__bu --VPAEVPDHMANIHLPAFQQNATVPRSGQLPAD-------QQEFTVTGA
Mb1086|M.bovis_AF2122/97 ----------MSVMNGREVARESRD---AQVFEFG-----TAPG-SAVVK
Rv1057|M.tuberculosis_H37Rv ----------MSVMNGREVARESRD---AQVFEFG-----TAPG-SAVVK
MMAR_4413|M.marinum_M ----------MSDVNGQQTGWNDRDGGDVQVFELG-----TAPDFPTSVE
MUL_0230|M.ulcerans_Agy99 ----------MSDVNGQQTGWNDRDGGDVQVFELG-----TAPDFPTSVE
MAV_1185|M.avium_104 MNPGANSPRPYRRLRHLATRADKHSKGGTSVSDANGPTARFGIDNTAVVK
:
Mvan_4699|M.vanbaalenii_PYR-1 ITLGRGPVGDIAAGQG--FVVMANAGDDSVSVIDPMTLAVVDTIAVDGQP
Mflv_2024|M.gilvum_PYR-GCK VRVQRGPIGDMATTDG--VVAIANPGDDSVSLLDPSTLTVVDTVVVDGEP
MSMEG_5308|M.smegmatis_MC2_155 VEVGRGPIADIAIDADRETIVVTNSAADCLTVINPYTLAPVGSVRLNGEP
TH_4450|M.thermoresistible__bu VAVD-GPVADLAINADGHALVAAHCGRDAVSLLDPARLNVQAAVPLGGDP
Mb1086|M.bovis_AF2122/97 IPVQGGPIGGIAISRDGSLLVVTNNGTDTVSVVGTDTCRVTQTVTSVNEP
Rv1057|M.tuberculosis_H37Rv IPVQGGPIGGIAISRDGSLLVVTNNGTDTVSVVGTDTCRVTQTVTSVNEP
MMAR_4413|M.marinum_M IAVANGPIGDIGISPDGRRLMVTNYGDNSVSVIDTDTCRVVETITGIDEA
MUL_0230|M.ulcerans_Agy99 IAVANGPIGDIGISPDGSRLMVTNYGDNSVSVIDTDTCRVVETITGLDEA
MAV_1185|M.avium_104 VPVHHGPISDIDVSPDGRRLLVTNYGRDTVSVIDTHTCRVASTIAGLSEP
: : **:..: . : :: . : ::::.. :: .:.
Mvan_4699|M.vanbaalenii_PYR-1 VAVTAS---DDRAYVC-VAAESHDAVCAIDLEAGTVVSTHPLASGVTALA
Mflv_2024|M.gilvum_PYR-GCK QAVAVA---DGRAYVG-IAAAGHDAVAVVDLDSRTLIKTYPLAFGVTALA
MSMEG_5308|M.smegmatis_MC2_155 FAVAAA---DDRAYVS-VVTAGHDAIKVVDTITGSVLAEYPLAMTVTALA
TH_4450|M.thermoresistible__bu FHVAVT---RDHAYVSTISLRNEDAVEVVELGSGTVVATHPQRATITALV
Mb1086|M.bovis_AF2122/97 FAIAMGNAEANRAYVS-TVSSAYDAIAVIDVATNTVLGTHPLALSVSDLT
Rv1057|M.tuberculosis_H37Rv FAIAMGNAEANRAYVS-TVSSAYDAIAVIDVATNTVLGTHPLALSVSDLT
MMAR_4413|M.marinum_M FAVTTGGPAANRAYVS-TVSTAYDAIAVIDVATNAVVSTHPLALSVSDLA
MUL_0230|M.ulcerans_Agy99 FAVTAGGPAANRAYVS-TVSTAYDAIAVIDVATNAVVSTHPLALSVSDLA
MAV_1185|M.avium_104 FAVAMSAADPNYAYVS-TATAAYDAIEVIDVVTNWRIATHRLAHSLSDLA
:: . *** **: .:: : : : :: *.
Mvan_4699|M.vanbaalenii_PYR-1 VSPDGKRVYAGRDAQDRVDVAVIDVTAE-RVGTIGIGSGPAANVDALSVD
Mflv_2024|M.gilvum_PYR-GCK VSPDGKRVYAGRCVRDRVDVAVIDVTAE-RVGTIDIGRGPAANIDAMSVD
MSMEG_5308|M.smegmatis_MC2_155 MSPDGKRVFVGRSGHDRIDVAVIDTAAE-RVGTIDLASGAGAGVDALRVD
TH_4450|M.thermoresistible__bu ASPDGKRVFVGRTDSERVDIAVIDTITG-EVDTVDAGWGPAINIDALHVV
Mb1086|M.bovis_AF2122/97 LSPDDKYLYVSRNGTRGADVAVLDTTTGALIDVVDVSQAPGTTTQCVRMS
Rv1057|M.tuberculosis_H37Rv LSPDDKYLYVSRNGTRGADVAVLDTTTGALIDVVDVSQAPGTTTQCVRMS
MMAR_4413|M.marinum_M ASSDGKYVYASRNGARGADIAVLDTATG-RVDVVDIATKPGLTTECVRVS
MUL_0230|M.ulcerans_Agy99 ASSDGKYVYASRNGARGADIAVLDTATG-RVDVVDIATKPGLTTECVRVS
MAV_1185|M.avium_104 VSPDGRYLYASRNAVRGADVAVLDTTTG-ELEVIELAAAAGTTTACVRVS
*.*.: ::..* *:**:*. : : .: . .. .: :
Mvan_4699|M.vanbaalenii_PYR-1 PSGKRLYVAVTDAVGSRLVIVDTETSLVHR--------------------
Mflv_2024|M.gilvum_PYR-GCK PGGKRMYLAVTDTEGSRLIVVDTETSRVSR--------------------
MSMEG_5308|M.smegmatis_MC2_155 ASGKRLYVATTDPRGSRMVTVNIETAQIES--------------------
TH_4450|M.thermoresistible__bu DNGRRLHAAVSDACSSYLVNINVETARVEG--------------------
Mb1086|M.bovis_AF2122/97 PDGSVLYVGANGPSGGLLVVITTRAQSD-----GGRIGSRSRSRQKSSKP
Rv1057|M.tuberculosis_H37Rv PDGSVLYVGANGPSGGLLVVITTRAQSD-----GGRIGSRSRSRQKSSKP
MMAR_4413|M.marinum_M ADGQRVYVGANGPSGGQLIVVATHAESELNPVRGGRARSRGRNGKGVAGS
MUL_0230|M.ulcerans_Agy99 ADGNRVDVGANGPSGGQLIVVATHAESELNPVRGGRARSRGRNGQGVAGS
MAV_1185|M.avium_104 ADGRRLFVGVNGPNGGGLAIIETRTRTD-----GRRVGGRSR--------
.* : ..... .. : : .:
Mvan_4699|M.vanbaalenii_PYR-1 -------------VLPIGAPIRDIAY--AGGAVYVLTSDRAVGGTVHVVD
Mflv_2024|M.gilvum_PYR-GCK -------------VVPIGSPIRDLGY--AAGALYALTSDRAVGGAIHVVD
MSMEG_5308|M.smegmatis_MC2_155 -------------TVWIGAPIRDLALG-ADGKALVLTSDRQRRGVVHIVD
TH_4450|M.thermoresistible__bu -------------AVGIGAPIRDVAVG-PDLIAYVLTSDRHRGGSVAKVD
Mb1086|M.bovis_AF2122/97 RGNQAAAGLRVVATIDIGSSVRDVALSPDGAIAYVASCGSDFGAVVDVID
Rv1057|M.tuberculosis_H37Rv RGNQAAAGLRVVATIDIGSSVRDVALSPDGAIAYVASCGSDFGAVVDVID
MMAR_4413|M.marinum_M VVKQFPRDLRIIDTIEIGSSVRDVAISPDGTTAYVASCGADFGAVIDVVD
MUL_0230|M.ulcerans_Agy99 VVQQFPRDLRIIDTIEIGSSVRDVAISPDGTTAYVASCGADFGAVIDVVD
MAV_1185|M.avium_104 ----------LVGTVELGLPVRDVALGNDGNTAYVASCGPVVGSVLDVVD
.: :* .:**:. . :.. . : :*
Mvan_4699|M.vanbaalenii_PYR-1 LNAMRVTDR---VVVGGAPTQLTMSADQTRAYIVDYDRVAVLCTLGLDVV
Mflv_2024|M.gilvum_PYR-GCK LATWKVTEA---TTLGGAPTQLVMSADQTRAYVVDYDRVAVVSTSSLEVS
MSMEG_5308|M.smegmatis_MC2_155 LSTAAVVGA---IQIGGAPTQLVLSPDATRAYVVDYDRVIVLCTLTNEIL
TH_4450|M.thermoresistible__bu LAHGRIVER---VDIGGAPTQLVLAADGSRAYIVDYDSVAVLCTVTNQVV
Mb1086|M.bovis_AF2122/97 TRTHQITSSRAISEIGGLVTRVSVSGDADRAYLVSEDRVTVLCTRTHDVI
Rv1057|M.tuberculosis_H37Rv TRTHQITSSRAISEIGGLVTRVSVSGDADRAYLVSEDRVTVLCTRTHDVI
MMAR_4413|M.marinum_M TRTRKITGTRKIEEIGGLITRLSIGGNGERAYLVSEDRVTVLSARSQDVI
MUL_0230|M.ulcerans_Agy99 TRTHKITGTRKIEEIGGLITRLSIGGNGERAYLVSEDRVTVLSARSQDVI
MAV_1185|M.avium_104 TRAATIVSTHKINEITGPLTRMTLSRDGERAYLISDDRVTVLGTRTQDVL
:. : * *:: :. : ***::. * * *: : ::
Mvan_4699|M.vanbaalenii_PYR-1 DSLAVDARPSCVTHPADSSRLYIADYTGAVSVFSVESSIETLYSQFLATD
Mflv_2024|M.gilvum_PYR-GCK DSLTAEARPSCVTQSADGTRLYIADYSGGVSVLSVESSIEALYPHFLATD
MSMEG_5308|M.smegmatis_MC2_155 GSIDVGMQPAAVAVRRDGARVYVADYSGQVNAFDVAAELPALYSRLVASE
TH_4450|M.thermoresistible__bu HAIDPGAAPSGVALSPDGGRLFIADHTGRITMFAVAGAQPLWPEAFLDTD
Mb1086|M.bovis_AF2122/97 GTIRTG-QPSCVVESPDGKYLYIADYSGTITRTAVASTIVSGTEQL-ALQ
Rv1057|M.tuberculosis_H37Rv GTIRTG-QPSCVVESPDGKYLYIADYSGTITRTAVASTIVSGTEQL-ALQ
MMAR_4413|M.marinum_M GTVRVN-QPSAAIESPGGDRLYVADYSGTVTMTLVDLG-VSAIDN--ALC
MUL_0230|M.ulcerans_Agy99 GTVRVN-QPSAAIESPGGDRLYVADYSGTVTMTLVDLG-VSAIDN--ALC
MAV_1185|M.avium_104 GEVRVTKHPSALVESPDGNYLYVADHAGVLSVARLPSGTAPAAPCSGADE
: *: .. :::**::* :. :
Mvan_4699|M.vanbaalenii_PYR-1 S--IALGVSRALQPTPA---
Mflv_2024|M.gilvum_PYR-GCK P--IAQTATRALQPAGV---
MSMEG_5308|M.smegmatis_MC2_155 PRQDATTVLPSLQTA-----
TH_4450|M.thermoresistible__bu P--IPAHRIPAEVA------
Mb1086|M.bovis_AF2122/97 RRGSMQWFSPELQQYAPALA
Rv1057|M.tuberculosis_H37Rv RRGSMQWFSPELQQYAPALA
MMAR_4413|M.marinum_M QGLSADWVMPELLQFEAALA
MUL_0230|M.ulcerans_Agy99 QGLSADWVMPELLQFEAALA
MAV_1185|M.avium_104 DDDATTGWLPELAPWEPVLA