For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLRVVQWATGGVGVAAIKGVLEHPELELVGCWVHSPGKAGRDVGEIVGLDPLGVTATDSIDDILALEADA VIYSPLIANPDEVAALLRSGKNVVTPVGWLYPSERSGASMREAALAGNATLHGTGIAPGGISEKFPLMLS AMSTGVTFVRAEEFSDLRTYEAPDVLRHVMGFGETPDKALTGPMQKMLDTGFIQAVRMCVDQLGFAADPK VLATQEIAVATARIDSPMGPIEPGQVAGRKFHWEALVDGEPVVRVTVNWLMGEENLDPAWSFGPEGQRYE MEVRGNPDFTVSVKGFQSEVGGEGPEYGVVGTAAHCVNSVPAVCAAPPGIATYLDLPLISGKAAPAKARA GI*
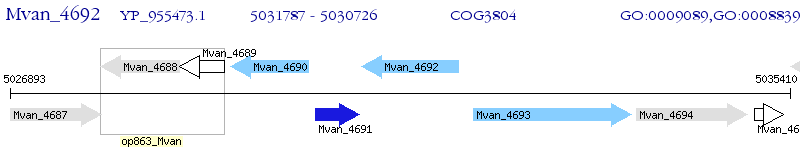
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4692 | - | - | 100% (353) | dihydrodipicolinate reductase |
| M. vanbaalenii PYR-1 | Mvan_4981 | - | 1e-57 | 39.02% (346) | dihydrodipicolinate reductase |
| M. vanbaalenii PYR-1 | Mvan_4980 | - | 8e-49 | 37.04% (351) | dihydrodipicolinate reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1088 | - | 1e-149 | 70.74% (352) | hypothetical protein Mb1088 |
| M. gilvum PYR-GCK | Mflv_2027 | - | 0.0 | 89.49% (352) | dihydrodipicolinate reductase |
| M. tuberculosis H37Rv | Rv1059 | - | 1e-149 | 70.74% (352) | hypothetical protein Rv1059 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4633 | - | 1e-42 | 34.84% (353) | hypothetical protein MAB_4633 |
| M. marinum M | MMAR_4411 | - | 1e-145 | 70.40% (348) | hypothetical protein MMAR_4411 |
| M. avium 104 | MAV_0353 | - | 1e-123 | 60.68% (351) | hypothetical protein MAV_0353 |
| M. smegmatis MC2 155 | MSMEG_5286 | dapB | 1e-168 | 81.50% (346) | dihydrodipicolinate reductase N-terminus domain-containing |
| M. thermoresistible (build 8) | TH_1493 | - | 1e-167 | 80.06% (346) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4353 | - | 1e-114 | 55.77% (355) | hypothetical protein MUL_4353 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1088|M.bovis_AF2122/97 --MTMSLRVIQWATGSVGVAAIKGVLQHPELELVGCWVHSAAKSGKDVGE
Rv1059|M.tuberculosis_H37Rv --MTMSLRVIQWATGSVGVAAIKGVLQHPELELVGCWVHSAAKSGKDVGE
MMAR_4411|M.marinum_M ----MGLRVVQWATGSVGVAAIKGVLEHPELELVGCWVYSEAKSGVDVGE
Mvan_4692|M.vanbaalenii_PYR-1 ----MTLRVVQWATGGVGVAAIKGVLEHPELELVGCWVHSPGKAGRDVGE
Mflv_2027|M.gilvum_PYR-GCK ----MALRVVQWATGGVGIAAIHGVLAHPELELVGCWVHSKAKAGRDVGD
MSMEG_5286|M.smegmatis_MC2_155 ----MALRVVQWATGGVGMAAIKGVLEHPALELVGCWVHSEGKAGRDVGE
TH_1493|M.thermoresistible__bu ----MTFRVVQWATGGVGIAAIKGVLEHPELELVGCWVHSPDKAGRDVGE
MAV_0353|M.avium_104 MSSPSKLRVIQWASGGVGKAAIQCVLNHPRLELAGCWVHSAEKNGVDVGR
MUL_4353|M.ulcerans_Agy99 MSGPAKLRVIQWATGGVGKAAIECILNHPQLELAGCWVHSAAKHGLDVGT
MAB_4633|M.abscessus_ATCC_1997 ---MTKTRVAVWGTGNVGQHALAGVITDPSMELVGVWVSGSNKAGKDAGA
** *.:*.** *: :: .* :**.* ** . * * *.*
Mb1088|M.bovis_AF2122/97 IIGSPPLG-VIATNSIDDVLALDADAVIYAPLLPS-----VDEVAALLRS
Rv1059|M.tuberculosis_H37Rv IIGSPPLG-VIATNSIDDVLALDADAVIYAPLLPS-----VDEVAALLRS
MMAR_4411|M.marinum_M IIGTPPLG-VITTNSVDEILALDADAVIYAPLMPS-----VDEVAALLHS
Mvan_4692|M.vanbaalenii_PYR-1 IVGLDPLG-VTATDSIDDILALEADAVIYSPLIAN-----PDEVAALLRS
Mflv_2027|M.gilvum_PYR-GCK IVGTAPLG-VVATDSIDEILSLDADAVIYSPLIAD-----TEEVVALLKS
MSMEG_5286|M.smegmatis_MC2_155 IIGTEPLG-VETTRDIDDILGLDADAVIYAPLLPN-----PEEVAALLRS
TH_1493|M.thermoresistible__bu IIGAEPLG-ITATNSIDDILALDADAVIYAPLLPD-----PDEVAALLRS
MAV_0353|M.avium_104 IIGTQELG-VTASTSVDEVLALDADCVVYSPLIPN-----DDEVIAILRS
MUL_4353|M.ulcerans_Agy99 IIGAARFG-VTATNDVDELLALDADCVMYSPQVAD-----EQDVKAILRS
MAB_4633|M.abscessus_ATCC_1997 LAGLDILTGITATTDAAQILALRPDCVVYTAMTDNRLMEALKDLTDLLSA
: * : : :: . ::*.* .*.*:*:. . .:: :* :
Mb1088|M.bovis_AF2122/97 GKNVVT--PLGWFYP----SEKEAAPLEVAAQAGNATLHGAGIGPGAVTE
Rv1059|M.tuberculosis_H37Rv GKNVVT--PLGWFYP----SEKEAAPLEVAAQAGNATLHGAGIGPGAVTE
MMAR_4411|M.marinum_M GKNVVT--PLGWFYP----TEKEAAPLEVAAQSGNATLHGAGIGPGAATE
Mvan_4692|M.vanbaalenii_PYR-1 GKNVVT--PVGWLYP----SERSGASMREAALAGNATLHGTGIAPGGISE
Mflv_2027|M.gilvum_PYR-GCK GKNVVT--PVGWLYP----SERSGAPLREAALAGDATLHGTGIAPGGISE
MSMEG_5286|M.smegmatis_MC2_155 GKNVVT--PVGWVYP----SERQGAPLREAALAGNATLHGTGIAPGGISE
TH_1493|M.thermoresistible__bu GKNVVT--PLGWFYP----TEKQAAPLRAAALEGGATLHGTGIAPGGISE
MAV_0353|M.avium_104 GKNVVT--PVGWVYPD--PGNPRHRAVADAALESGVTLHGSGIHPGGITE
MUL_4353|M.ulcerans_Agy99 GKNVFT--PVGWVYPD--LSKPRVKAVADAAVAGGVTLHGSGIHPGGITE
MAB_4633|M.abscessus_ATCC_1997 GINVAASAPVFLQYPWGVLPENMLTPIENAAREGNSSIFAGGIDPGFAND
* ** : *: ** : .: ** .. ::.. ** ** .:
Mb1088|M.bovis_AF2122/97 LFPLLLSVMSTGVTFVRSEEFSDLRSYGAPDVLRYVMGFGGTPDSALTGP
Rv1059|M.tuberculosis_H37Rv LFPLLLSVMSTGVTFVRSEEFSDLRSYGAPDVLRYVMGFGGTPDSALTGP
MMAR_4411|M.marinum_M LFPLLLSVMSTGVTFVRSEEFSDLRTYGAPDVLRYVMGFGGTPDSALTGP
Mvan_4692|M.vanbaalenii_PYR-1 KFPLMLSAMSTGVTFVRAEEFSDLRTYEAPDVLRHVMGFGETPDKALTGP
Mflv_2027|M.gilvum_PYR-GCK KFPLMLSAMSTGVTFVRAEEYSDLRTYEAPDVLRHVMGFGETPDKALTGP
MSMEG_5286|M.smegmatis_MC2_155 KFPLLFSVFSTGVTFVRAEEFSDLRTYDAPDVLRHVMGFGEVPDKALSGP
TH_1493|M.thermoresistible__bu KLPLMFSALATGVTFVRAEEFSDLRTYDAPDVVRHVMGFGDTPEKALSGP
MAV_0353|M.avium_104 RFPLMVSSLSSAVTHVRAEEFSDIRTYNAPDVVRHIMGFGGTPEEATGGP
MUL_4353|M.ulcerans_Agy99 RFPLMVSSLSSAITHVRAEEFSDIRTYNAPDVVRDIMGFGVPHAQAMQGP
MAB_4633|M.abscessus_ATCC_1997 LLPLALMGTCQRIDQVRCIEIVDYATYDSATVMFDVMGFGKPMEETPMLL
:** . . : **. * * :* :. *: :**** .:
Mb1088|M.bovis_AF2122/97 MQKILDGGFLQSVRLCVDRLGFAADPQIRTSQEVAVATAPIDSPIGVIEP
Rv1059|M.tuberculosis_H37Rv MQKILDGGFLQSVRLCVDRLGFAADPQIRTSQEVAVATAPIDSPIGVIEP
MMAR_4411|M.marinum_M MQKLLDGGFMQSVRLCVDRLGFAADPQIRTSQEVAVATSPIESPIGVIEP
Mvan_4692|M.vanbaalenii_PYR-1 MQKMLDTGFIQAVRMCVDQLGFAADPKVLATQEIAVATARIDSPMGPIEP
Mflv_2027|M.gilvum_PYR-GCK MQKMLDSGFIQAVRMVVDQLGFAADPKVRATQEIAVATAPIDSPMGQILP
MSMEG_5286|M.smegmatis_MC2_155 MQKLLDGGFMQAVKMCVDQFGFAADPKIRSRQEIAVATAPIDSPLGAIQP
TH_1493|M.thermoresistible__bu MQKLLDGGFIQAVRMCVDKIGFAADPQVRSRQEIAVATAPIESPIGVIEP
MAV_0353|M.avium_104 MAGLLDGGFKQSVRMIADHMGFRIDPNIRTIQDVAVATADIDYDPFPITA
MUL_4353|M.ulcerans_Agy99 MAALLESGFKMSVRMIANHMGFHIDPMIRATHEIAVATSDIASDVLPIAA
MAB_4633|M.abscessus_ATCC_1997 QPGVLSLAWGSVVRQLAAGLGVELD-EVTEVYEKLSAPEDFDISSGHIPA
:*. .: *: . :*. * : : *. : * .
Mb1088|M.bovis_AF2122/97 GQVAGRRFHWEALVEDTVVVQIAVNWLMGSENLDPPWSFGP-AGERYEIE
Rv1059|M.tuberculosis_H37Rv GQVAGRRFHWEALVEDTVVVQIAVNWLMGSENLDPPWSFGP-AGERYEIE
MMAR_4411|M.marinum_M GQVAGRRFHWEAVVGDTAVVEITVNWLMGEENLDPPWSFGP-AGERYEIE
Mvan_4692|M.vanbaalenii_PYR-1 GQVAGRKFHWEALVDGEPVVRVTVNWLMGEENLDPAWSFGP-EGQRYEME
Mflv_2027|M.gilvum_PYR-GCK GQVAGRKFHWEALVGGQPVVRVTVNWLMGEENLEPAWSFGP-EGQRYEME
MSMEG_5286|M.smegmatis_MC2_155 GQVAGRRFHWEALRGEQVVVRVTVNWLMGEENLDPPWTFGP-QGQRYEMQ
TH_1493|M.thermoresistible__bu GQVAGRRFHWEALAGDEVVVRVTVNWLMGEEDLDPPWDFGP-AGQRYEIE
MAV_0353|M.avium_104 GTVAARRFRWQALVDGEPVITAAVNWLMGEQNLDPAWDFGG-RGERFEVE
MUL_4353|M.ulcerans_Agy99 DTVAGRKFRWQTLADGKPVITAAVNWLMGQADMHPDWSFGE-RGERFEIE
MAB_4633|M.abscessus_ATCC_1997 GTMAALHFEVRGIKDGR-AVTVLEHFTRLREDLAPNWPQPAHEGGTYRVE
. :*. :*. . : .: :: :: * * * :.::
Mb1088|M.bovis_AF2122/97 VRGSPDTCVTIKGWQPQTVAAGLKSNPGIVATAAHCVNAIPATCAAPAGI
Rv1059|M.tuberculosis_H37Rv VRGSPDTCVTIKGWQPQTVAAGLKSNPGIVATAAHCVNAIPATCAAPAGI
MMAR_4411|M.marinum_M VRGHPDAFVTIKGWQPDSVAAGLQSNPGIVATAAHCVNAVPATCAAPAGI
Mvan_4692|M.vanbaalenii_PYR-1 VRGNPDFTVSVKGFQSEVGGEGPE--YGVVGTAAHCVNSVPAVCAAPPGI
Mflv_2027|M.gilvum_PYR-GCK VRGNPDFTVSVKGFQSDIGGEGPE--YGVVGTAAHCVNSVPAVCAAAPGI
MSMEG_5286|M.smegmatis_MC2_155 VCGNPDFDVVIKGFQGEVGETGPE--PGVVATAAHCVNSIPAVCAAEPGI
TH_1493|M.thermoresistible__bu VRGNPDFTVTINGFQSEPGDHGPE--AGIVATAAHCVNSVPAVCAADPGL
MAV_0353|M.avium_104 ITGDPDVSLTFKGLQPETIAEGLVKNPGVVVTANHCVNAIPDVCAAAPGI
MUL_4353|M.ulcerans_Agy99 ITGNPSVSLVFKGLQPTSITEGLQRNPGVVATANHCANAIPVVCAADPGI
MAB_4633|M.abscessus_ATCC_1997 ITGEPSYLLDLGSFS----ARGDHNYANLIATAMRVVNAVPAVVAAEAGI
: * *. : . . . * .:: ** : .*::* . ** .*:
Mb1088|M.bovis_AF2122/97 QSFFDLPLITGRAAPGLAR---
Rv1059|M.tuberculosis_H37Rv QSFFDLPLITGRAAPGLAR---
MMAR_4411|M.marinum_M QSFFDLPLITGRAAPELSN---
Mvan_4692|M.vanbaalenii_PYR-1 ATYLDLPLISGKAAPAKARAGI
Mflv_2027|M.gilvum_PYR-GCK ATYLDLPLISGKAAPDRARTGS
MSMEG_5286|M.smegmatis_MC2_155 ATYLDLPLFSAKAAPHLR----
TH_1493|M.thermoresistible__bu LTYLDMPLISGRAAPRLT----
MAV_0353|M.avium_104 KTYLDLPLFAGRPAPGLATS--
MUL_4353|M.ulcerans_Agy99 ATYLDLPLFAGRPARASRGLER
MAB_4633|M.abscessus_ATCC_1997 RTTLDLPLITGKGRAV------
: :*:**::.: