For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTLLAQTETIGNPVANIGIFSLFVVVTMIVVIRASKRNATADEFFTGGRGFSGPQNGIAIAGDYLSAAS FLGIAGAIAVYGYDGFLYSIGFLVAWLVALLLVAELLRNTGRFTMADVLSFRLKQRPVRLAAATNTLTVS LFYLLAQMAGAGGLVALLLDINSRTGQSVVIAVVGVLMIVYVLVGGMKGTTWVQIIKAVLLIAGAGFMTV MVLAKFGMNFSEILGSAQSAISGSTTTGVAGRDVLAPGAQYGGSLTSQINFISLAIALVLGTAGLPHVLM RFYTVPTAKEARRSVVWAIALIGAFYLFTLVLGYGAAALVGPDRILGAAGGVNSAAPLLAFELGGVVLLG VISAVAFATILAVVAGLTITASASFAHDIYASVMKSHKVTEAEQVRVSRITAVVLGVLAIGLGILANGQN IAFLVALAFAVAAAANLPTIIYSLYWRRFNTRGALWSMYGGLISTIVLIVFSPAVSGSKTAMIPGADFAW FPLANPGIVSIPLAFILGIVGTLTSPDDEDPKVAAEMEVRSLTGIGAEKAVAH
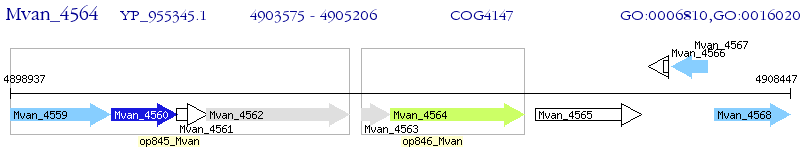
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4564 | - | - | 100% (543) | SSS family solute/sodium (Na+) symporter |
| M. vanbaalenii PYR-1 | Mvan_4562 | - | 6e-54 | 29.92% (595) | Na+/solute symporter |
| M. vanbaalenii PYR-1 | Mvan_0815 | - | 2e-11 | 22.32% (475) | Na+/solute symporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2139 | - | 0.0 | 97.05% (543) | SSS family solute/sodium (Na+) symporter |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4639 | - | 1e-13 | 20.41% (534) | putative Na+/solute symporter |
| M. marinum M | MMAR_3960 | - | 7e-08 | 25.92% (355) | hypothetical protein MMAR_3960 |
| M. avium 104 | MAV_1539 | - | 7e-05 | 27.49% (291) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_5166 | - | 0.0 | 92.76% (539) | Na+/solute symporter |
| M. thermoresistible (build 8) | TH_3825 | - | 3e-16 | 21.44% (471) | PUTATIVE putative Na+/solute symporter |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4564|M.vanbaalenii_PYR-1 -MTTLLAQTETIGNPVANIGIFSLFVVVTMIVVIRASKRNATADEFFTGG
Mflv_2139|M.gilvum_PYR-GCK -MTTLLAQSETVGNPLANIGIFSLFVVVTMIVVIRASKRNATADEFFTGG
MSMEG_5166|M.smegmatis_MC2_155 -MNVTLHAAETVGNPAANIGIFSLFVVVTLFVVIRASKRNATATEFFTGG
TH_3825|M.thermoresistible__bu ---------------MILTGVAVFLLVLTVSGVVSARRVRGDCANYLVAG
MAB_4639|M.abscessus_ATCC_1997 -----MTGSLRLDAGPVDYVLIAIYFVFVLGIGLLARRGVSSSIDFFLSG
MMAR_3960|M.marinum_M MTESQHAVISRLSAEFDSLARQMARVSGELSQLDRLISDSVGAPQPAAAA
MAV_1539|M.avium_104 ----------------------MVMPNGPVAHQAGTRGYRRMTAALCGAG
: ..
Mvan_4564|M.vanbaalenii_PYR-1 RGFSGPQNGIAIAGDYLSAASFLGIAGAIAVYG-YDGFLYSIGFLVAWLV
Mflv_2139|M.gilvum_PYR-GCK RGFSGPQNGIAIAGDYLSAASFLGIAGAIAVYG-YDGFLYSIGFLVAWLV
MSMEG_5166|M.smegmatis_MC2_155 RGFSGPQNGIAIAGDYLSAASFLGIAGAIAVYG-YDGFLYSIGFLVAWLV
TH_3825|M.thermoresistible__bu RALPAVLVAAVLIAQPVDSNATIGNADLSAAFGFWAGAALPIGLALCLLL
MAB_4639|M.abscessus_ATCC_1997 RSLPAWVTGLAFVSANLGAVEIMGMSANGAQFGMVGMHYFWIGAVPAILF
MMAR_3960|M.marinum_M PPQPQPQPVAPYWYPYWQQWAPVAPAPAAAPPRQYAPPPTARPAPQGPPA
MAV_1539|M.avium_104 LASFAAMYCTQALLPALSAHHRIGPATAALTVS------LTTGALALSII
:. :
Mvan_4564|M.vanbaalenii_PYR-1 ALLLVAELLRNTGRFTMADVLSFRLKQRPVRLAAATNTLTVSLFYLLAQM
Mflv_2139|M.gilvum_PYR-GCK ALLLVAELLRNTGRFTMADVLSFRLKQRPVRLAAATNTLTVSLFYLLAQM
MSMEG_5166|M.smegmatis_MC2_155 ALLLVAELLRNTGKFTMADVLSFRLKQRPVRMAAATSTLTVSLFYLLAQM
TH_3825|M.thermoresistible__bu MGLFFARRLRAAQVVTLPEYFGNRFG-RGVEVASSALTVGSFAILLAGNL
MAB_4639|M.abscessus_ATCC_1997 LGIVMMPFFYGSQVRSVPEYMLHRFG-VGAHLVNATGFAVAQVLIAGVNL
MMAR_3960|M.marinum_M PAPAAAPPVSAPRRPAESTESGWIGKLLAVAGVAVTLTGVALLLVLAAQA
MAV_1539|M.avium_104 PASVLSERYGRTRVMLISGVASSVIGLLLPFSPSLGVLVFGRAVQGVALA
. . .
Mvan_4564|M.vanbaalenii_PYR-1 AGAGGLVALLLDINSRTGQSVVIAVVGVLMIVYVLVGGMKGTTWVQIIKA
Mflv_2139|M.gilvum_PYR-GCK AGAGGLVALLLDINSRAGQSVVIAVVGVLMIVYVLVGGMKGTTWVQIIKA
MSMEG_5166|M.smegmatis_MC2_155 AGAGGLVALLLDVNSRSGQSLVIAVVGVLMIVYVLVGGMKGTTWVQIIKA
TH_3825|M.thermoresistible__bu VAIGFLLERFLGVSYTIG----ILVTVPLALLYTITGGMFASVYTGVAQV
MAB_4639|M.abscessus_ATCC_1997 YLLGTIVNVLLGWPLWVS----VIAAAAIVLCYITLGGLSAAIYNEVLQF
MMAR_3960|M.marinum_M GLLRPEIRVAGGTALSLA---LVGVAVRLRGRPGGRIGAIALAATGIATA
MAV_1539|M.avium_104 GIPAVAMALLAEEVDASSLGSAMGRYIAGTTIGGLAGRIVPSVVVQVGTW
: . : :
Mvan_4564|M.vanbaalenii_PYR-1 VLLIAGAGFMTVMVLAKFG--MNFSEILGS--AQSAISGSTTTGVAGRDV
Mflv_2139|M.gilvum_PYR-GCK VLLIAGAGFMTVMVLAKFG--LNFSEILGS--AQSAISGATTAGVADRDV
MSMEG_5166|M.smegmatis_MC2_155 VLLIAGAGLMTVMVLVKFG--INLSDILGS--AQTAISESTTQGVASRDV
TH_3825|M.thermoresistible__bu VVNIVGVFALVVWVAATHG--FTAPDGMGAG-ALSQLTSSESGAVIN---
MAB_4639|M.abscessus_ATCC_1997 FVIVAALLPLTIVGLHQVGGWQGLQDKVSAG-QMSSWPGNELSGFSDNLL
MMAR_3960|M.marinum_M YLDVIAATTIYQWVPAPAG--LVLAALVGG--AGLALARRWDSEHLGLLV
MAV_1539|M.avium_104 RVALLACSLITLAGTAVFAVLVPRSRFFTPKPASVRAALRNLAGHLRNPV
: : . : . . .
Mvan_4564|M.vanbaalenii_PYR-1 LAPGAQYGGSLTSQINFISLAIALVLGTAGLPHVLMRFYTVPTAK-----
Mflv_2139|M.gilvum_PYR-GCK LAPGAQYGGSLTSQINFISLALALVLGTAGLPHVLMRFYTVPTAK-----
MSMEG_5166|M.smegmatis_MC2_155 LAPGAQYGGSFTSQINFISLALALVLGTAGLPHVLMRFYTVPTAK-----
TH_3825|M.thermoresistible__bu -----------------WATIIALGLGNLVAIDLMQRVFSARDPG-----
MAB_4639|M.abscessus_ATCC_1997 SVIGIVFGLGFVLSFGYWTTNFVEVQRALAS-ESLSAARRTPIIG-----
MMAR_3960|M.marinum_M LVPLIGLAPVLTRGADLLLVSFMLALSAAALPVQLGKDWVWLHAARVAAP
MAV_1539|M.avium_104 LAK-------------LFAVGFVLMGGFVTVYNYLGYRLAARPFG-----
: :
Mvan_4564|M.vanbaalenii_PYR-1 --------------------EARRSVVWAIALIGAFYLFT---LVLGYGA
Mflv_2139|M.gilvum_PYR-GCK --------------------EARRSVVWAIALIGAFYLFT---LVLGYGA
MSMEG_5166|M.smegmatis_MC2_155 --------------------EARRSVVWAIALIGAFYLFT---LVLGYGA
TH_3825|M.thermoresistible__bu --------------------AAQR---ACFAAAGGILLLC---LPLSFVA
MAB_4639|M.abscessus_ATCC_1997 --------------------AFPKMFIPFIVVIPGMICAV---LVPQMVA
MMAR_3960|M.marinum_M TVPLLIALVAAGKHDNTWLLGAACGVAAILAIVSGLIVLPGGKNAAALAV
MAV_1539|M.avium_104 ---------------------------LAPSVVGLLFLLY----------
:
Mvan_4564|M.vanbaalenii_PYR-1 AALVGPDRILGAAGGVNSAAPLLAFELGGVVLLGVISAV-----AFATIL
Mflv_2139|M.gilvum_PYR-GCK AALVGPDRILGAAGGVNSAAPLLAFELGGVILLGVISAV-----AFATIL
MSMEG_5166|M.smegmatis_MC2_155 AALVGPDRILGAAGGVNSAAPLLAFELGGVILLGIISAV-----AFATIL
TH_3825|M.thermoresistible__bu LAAVG---IVGDSAGDGPILYALLGEYAPAWLAIIVISG-----LLAATL
MAB_4639|M.abscessus_ATCC_1997 YKAGQP--TSDPNLTYSNSLLYLMKEVLPNGVLGVAITG-----LLASFM
MMAR_3960|M.marinum_M LTVVGTLPVLAAAVAVDRVFAALLAGTLAAALLAIVLVGSELPTIFTRVL
MAV_1539|M.avium_104 -LVGTGTSVVAGRLADRRGRPLVLGAALPIAVAGLLLTVP------PSLA
: : : .
Mvan_4564|M.vanbaalenii_PYR-1 AVVAGLTITASASFAHDIY-----ASVMKSHKVTEAEQVRVSRITAVVLG
Mflv_2139|M.gilvum_PYR-GCK AVVAGLTITASASFAHDIY-----ASVLKSHKVTEAEQVKVSRITAVVLG
MSMEG_5166|M.smegmatis_MC2_155 AVVAGLTITASASFAHDVY-----ASVLKSHKVTEDEQVRVSRITAVVLG
TH_3825|M.thermoresistible__bu TTVSGILLSTASVLVRNVLRI--DTGVLAPPAAMTPRMLRATRWATLPMA
MAB_4639|M.abscessus_ATCC_1997 AGMAANVSAFNTVFSYDLW------QRYIRKDRPDGYYLRIGRIATVGAT
MMAR_3960|M.marinum_M AAWSAISALVAVTVAFDGHVEGPILLALAIVVAIAGRHDRMALWAAAGFG
MAV_1539|M.avium_104 AIVAGVGVFTGGFFAAHTVASG-WVGAVAQRDRAEASALYLFSYYLGGSV
: :. . .
Mvan_4564|M.vanbaalenii_PYR-1 VLAIGLGILANGQ-NIAFLVALAFAVAAAANLPTIIYSLYWRRFNTRGA-
Mflv_2139|M.gilvum_PYR-GCK ILAIGLGILANGQ-NIAFLVALAFAVAAAANLPTIIYSLYWKRFNTRGA-
MSMEG_5166|M.smegmatis_MC2_155 VLAIGLGILANGQ-NIAFLVALAFAVAAAANLPTIVYSLYWKRFNTRGA-
TH_3825|M.thermoresistible__bu ALAAVVALRVP---QTGILLTLAFDLLLASLVVPFILGLYWSRGGAAAA-
MAB_4639|M.abscessus_ATCC_1997 VIAIGTAFIAANYNNVMNYLQTLFAFFNAPLFATFILGMFWRRMTATAG-
MMAR_3960|M.marinum_M IVGLGLYYSYAPLHALLTATVMPTSIAVSTLAASLLSIAFVVAMSRACAG
MAV_1539|M.avium_104 AGAFGGVLYGVGGWSATVCFVVVLLMAGAALVALLVRDNGFRIG------
. . :. ::
Mvan_4564|M.vanbaalenii_PYR-1 ----------LWSMYGGLISTIVLIVFSPA-------------VSGSKTA
Mflv_2139|M.gilvum_PYR-GCK ----------LWSMYGGLISTIVLIVFSPA-------------VSGSKTA
MSMEG_5166|M.smegmatis_MC2_155 ----------LWSMYGGLISTIVLIVFSPA-------------VSGSKTA
TH_3825|M.thermoresistible__bu ----------AAAVVVGIGVRLTYFVLAPT-------------IYG----
MAB_4639|M.abscessus_ATCC_1997 ----------WAGLVSGTFAAVGIWSLFETGVYTLAGQGSNFIEAGVAFT
MMAR_3960|M.marinum_M IGGQNRDIRRILAAVGGVLVIYAVTAFTVTAG---------VLIAGTEAG
MAV_1539|M.avium_104 -----------RRVVAGVASVK----------------------------
*
Mvan_4564|M.vanbaalenii_PYR-1 MIP-GADFAWFPLANPGIVSIPLAFILGIVGTLTSPDDEDP---KVAAEM
Mflv_2139|M.gilvum_PYR-GCK MIP-GADFAFFPLANPGIVSIPLAFILGIVGTLTSPDDEDP---KVAAEM
MSMEG_5166|M.smegmatis_MC2_155 MIP-GADFDFFPLANPGIVSIPLAFILGIVGTLTSPDDEDP---AIAAEM
TH_3825|M.thermoresistible__bu -----VDNTLLYLPNDWVSAX------XRGGGPSGVRDTPS---DTPREM
MAB_4639|M.abscessus_ATCC_1997 VDI-VVSVAVSWTTEPKPAAELAGLVYSETPHAFSSDEPASGWFRQPVKL
MMAR_3960|M.marinum_M FLAGHMAATICWIAVAAALFVYALRVADRERRTAPIGAGLALTGAATAKL
MAV_1539|M.avium_104 --------------------------------------------------
Mvan_4564|M.vanbaalenii_PYR-1 EVRSLTGIGAEKAVAH---------------------
Mflv_2139|M.gilvum_PYR-GCK EVRSLTGIGAEKAVSH---------------------
MSMEG_5166|M.smegmatis_MC2_155 EVRSLTGVGAEKAISH---------------------
TH_3825|M.thermoresistible__bu HPRNALA------------------------------
MAB_4639|M.abscessus_ATCC_1997 AMTALILVVALNLIFH---------------------
MMAR_3960|M.marinum_M FLFDLATLDGIFRVAAFIVVGLVLLAMGTGYARSLAR
MAV_1539|M.avium_104 -------------------------------------