For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AEIVLDRVTKSYADGTAAVKDLSITVADGEFVTLVGPSGCGKSTTLNMIAGLEEITSGELRIDGERVNER STKDRDIAMVFQSYALYPHMTVRQNIGFPLTLAKLSRQEIAEKVEQTAKTLDLTDLLDRKPDQLSGGQRQ RVAMGRAIVRNPKAFLMDEPLSNLDAKLRVQMRGEIARLQKRLGTTTVYVTHDQTEAMTLGDRVVVMLGG AVQQIGTPDELYRGPANLFVAGFIGSPAMNFFPAALTEIGVRLPFGEIPLTGQVYERLKGHRTCDDLIAG IRPEHLEDAAVIDAYARIGALVFDVTVEMVESLGADKYVYFGVDGPGAEAVQLTEVAAESTVGENEFVAR VPAQSGAAVGRTLQLALNPDNVLLFDPRTGANLSLPAVGA*
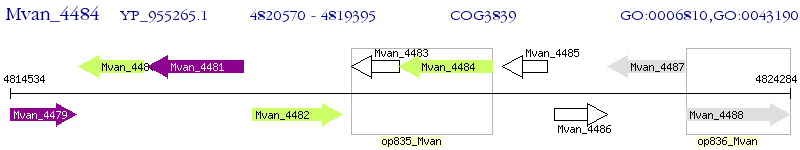
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4484 | - | - | 100% (391) | ABC transporter-related protein |
| M. vanbaalenii PYR-1 | Mvan_2541 | - | e-106 | 53.35% (388) | ABC transporter-related protein |
| M. vanbaalenii PYR-1 | Mvan_2621 | - | 2e-79 | 46.25% (387) | ABC transporter-related protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1270 | sugC | 1e-158 | 71.28% (390) | sugar-transport ATP-binding protein ABC transporter SugC |
| M. gilvum PYR-GCK | Mflv_3854 | - | 1e-104 | 52.84% (388) | ABC transporter related |
| M. tuberculosis H37Rv | Rv1238 | sugC | 1e-158 | 71.28% (390) | sugar-transport ATP-binding protein ABC transporter SugC |
| M. leprae Br4923 | MLBr_01089 | - | 1e-149 | 68.56% (388) | putative ABC-transport protein, ATP-binding component |
| M. abscessus ATCC 19977 | MAB_1375 | - | 1e-151 | 68.56% (388) | sugar ABC transporter, ATP-binding protein SugC |
| M. marinum M | MMAR_4203 | sugC | 1e-154 | 71.54% (390) | sugar-transport ATP-binding protein ABC transporter SugC |
| M. avium 104 | MAV_1377 | sugC | 1e-161 | 73.58% (386) | ABC transporter, ATP-binding protein SugC |
| M. smegmatis MC2 155 | MSMEG_5058 | - | 1e-159 | 72.75% (389) | ABC transporter, ATP-binding protein SugC |
| M. thermoresistible (build 8) | TH_4034 | - | 1e-165 | 74.94% (391) | PUTATIVE ABC transporter, ATP-binding protein SugC |
| M. ulcerans Agy99 | MUL_2252 | - | 5e-90 | 51.90% (343) | sugar-transport ATP-binding protein ABC transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4484|M.vanbaalenii_PYR-1 MAEIVLDRVTKSYADGT----AAVKDLSITVADGEFVTLVGPSGCGKSTT
TH_4034|M.thermoresistible__bu MAEIVLDKVTKSYPDGAGGVRHAVKDLSLTIADGEFIILVGPSGCGKSTT
MSMEG_5058|M.smegmatis_MC2_155 MAEIVLDRVTKSYPDGAGGVRAAVKEFSMTIADGEFIILVGPSGCGKSTT
Mb1270|M.bovis_AF2122/97 MAEIVLDHVNKSYPDGH----TAVRDLNLTIADGEFLILVGPSGCGKTTT
Rv1238|M.tuberculosis_H37Rv MAEIVLDHVNKSYPDGH----TAVRDLNLTIADGEFLILVGPSGCGKTTT
MMAR_4203|M.marinum_M MAEIVLEHVNKSYPDGH----TAVRDLNITISDGEFLILVGPSGCGKTTT
MAV_1377|M.avium_104 MAEIVLDHVSKSYPDGA----TAVRDLNLTIADGEFLILVGPSGCGKTTT
MLBr_01089|M.leprae_Br4923 MAEIVLEHVNKNYPNGA----TAVHDLSITVADGEFLILIGPSGCGKTTT
MAB_1375|M.abscessus_ATCC_1997 MAEIVLNQVSKTYTDGS----AAVRDVSLNIADGEFIILVGPSGCGKSTT
Mflv_3854|M.gilvum_PYR-GCK MAAITMRNIVKKYGDGY----PAVNDVSLDIADGEFVILVGPSGCGKSTL
MUL_2252|M.ulcerans_Agy99 MASVSFEGATRRYPGADR---PAVDSLDLVVDDGEFVVLVGPSGCGKTTS
** : : : * .. ** ...: : ****: *:*******:*
Mvan_4484|M.vanbaalenii_PYR-1 LNMIAGLEEITSGELRIDGERVNERSTKDRDIAMVFQSYALYPHMTVRQN
TH_4034|M.thermoresistible__bu LNMIAGLEDITSGEIRIDGERVNEVAPKDRDIAMVFQSYALYPHMTVRQN
MSMEG_5058|M.smegmatis_MC2_155 LNMIAGLEEITSGELRIGGERVNEKAPKDRDIAMVFQSYALYPHMTVRQN
Mb1270|M.bovis_AF2122/97 LNMIAGLEDISSGELRIAGERVNEKAPKDRDIAMVFQSYALYPHMTVRQN
Rv1238|M.tuberculosis_H37Rv LNMIAGLEDISSGELRIAGERVNEKAPKDRDIAMVFQSYALYPHMTVRQN
MMAR_4203|M.marinum_M LNMIAGLEDISSGELRIGGERVNEKAPKDRDIAMVFQSYALYPHMTVRQN
MAV_1377|M.avium_104 LNMIAGLEDISSGELRIGGERVNEKAPKDRDIAMVFQSYALYPHMTVRQN
MLBr_01089|M.leprae_Br4923 LNMIAGLEDISSGELRIDGDRVNEKAPKDRDIAMVFQSYALYPHMTVRQN
MAB_1375|M.abscessus_ATCC_1997 LNMIAGLEDISSGELTIAGERVNEKAPKDRDIAMVFQSYALYPHMTVRQN
Mflv_3854|M.gilvum_PYR-GCK LRMIVGLEDITSGDMMIGDKRVNDMAPRDRNLAMVFQNYALYPHLTVFEN
MUL_2252|M.ulcerans_Agy99 LRMLAGLEKVDAGRIRIAERDVTNVDPKSRDVAMVFQNYALYPHMTVAQN
*.*:.***.: :* : * *.: .:.*::*****.******:** :*
Mvan_4484|M.vanbaalenii_PYR-1 IGFPLTLA-KLSRQEIAEKVEQTAKTLDLTDLLDRKPDQLSGGQRQRVAM
TH_4034|M.thermoresistible__bu IAFPLTLA-KMKKADIARKVEETAKILDLTELLDRKPAQLSGGQRQRVAM
MSMEG_5058|M.smegmatis_MC2_155 IAFPLTLA-KVPKAEIAAKVEETAKILDLSELLDRKPGQLSGGQRQRVAM
Mb1270|M.bovis_AF2122/97 IAFPLTLA-KMRKADIAQKVSETAKILDLTNLLDRKPSQLSGGQRQRVAM
Rv1238|M.tuberculosis_H37Rv IAFPLTLA-KMRKADIAQKVSETAKILDLTNLLDRKPSQLSGGQRQRVAM
MMAR_4203|M.marinum_M IAFPLTLA-KMKKAAIAQKVEETAKILDLTSFLDRKPSQLSGGQRQRVAM
MAV_1377|M.avium_104 IAFPLTLA-KMKKPEIAQKVAETAKILDLTDFLDRKPSQLSGGQRQRVAM
MLBr_01089|M.leprae_Br4923 IAFPLMLA-KVKKAEIAQKVSETAQILDLTDLLDRKPSQLSGGQRQRVAM
MAB_1375|M.abscessus_ATCC_1997 IAFPLTLA-KMSKDDINAKVAEAAKILDLTDYLDRKPANLSGGQRQRVAM
Mflv_3854|M.gilvum_PYR-GCK IAFPLRLAGKLSDDEIRQRVNDAATTLELGEHLDRKPANLSGGQRQRVAM
MUL_2252|M.ulcerans_Agy99 MGFALKVA-KTPKAEIRDRVLQAAKLLDLEAYLDRKPKDLSGGQRQRVAM
:.*.* :* * * :* ::* *:* ***** :***********
Mvan_4484|M.vanbaalenii_PYR-1 GRAIVRNPKAFLMDEPLSNLDAKLRVQMRGEIARLQKRLGTTTVYVTHDQ
TH_4034|M.thermoresistible__bu GRAIVRQPKAFLMDEPLSNLDAKLRVQMRSEIARLQSRLRTTTVYVTHDQ
MSMEG_5058|M.smegmatis_MC2_155 GRAIVRSPKAFLMDEPLSNLDAKLRVQMRAEISRLQDRLGTTTVYVTHDQ
Mb1270|M.bovis_AF2122/97 GRAIVRHPKAFLMDEPLSNLDAKLRVQMRGEIAQLQRRLGTTTVYVTHDQ
Rv1238|M.tuberculosis_H37Rv GRAIVRHPKAFLMDEPLSNLDAKLRVQMRGEIAQLQRRLGTTTVYVTHDQ
MMAR_4203|M.marinum_M GRAIVRHPKAFLMDEPLSNLDAKLRVQMRGEIARLQKRLGTTTVYVTHDQ
MAV_1377|M.avium_104 GRAIVRHPKAFLMDEPLSNLDAKLRVQMRGEIARLQKRLGTTTVYVTHDQ
MLBr_01089|M.leprae_Br4923 GRAIVRHPKAFLMDEPLSNLDAKLRVRTRGEIARLQRRLGATTVYVTHDQ
MAB_1375|M.abscessus_ATCC_1997 GRAIVRSPKAFLMDEPLSNLDAKLRVQMRTEIARLQKRLGTTTVYVTHDQ
Mflv_3854|M.gilvum_PYR-GCK GRAIVREADAFLFDEPLSNLDAKLRGQMRTEILRLQRKLGVTTVYVTHDQ
MUL_2252|M.ulcerans_Agy99 GRAIVRRPQVFLMDEPLSNLDTKLRVQTRNQIAGLQRQLGTTTVYVTHDQ
****** ...**:********:*** : * :* ** :* .*********
Mvan_4484|M.vanbaalenii_PYR-1 TEAMTLGDRVVVMLGGAVQQIGTPDELYRGPANLFVAGFIGSPAMNFFPA
TH_4034|M.thermoresistible__bu TEAMTLGDRVVVMLDGVAQQIGTPDELYTQPANLFVAGFIGSPGMNFFPA
MSMEG_5058|M.smegmatis_MC2_155 TEAMTLGDRVVVMLAGEVQQIGTPDELYSSPANLFVAGFIGSPAMNFFPA
Mb1270|M.bovis_AF2122/97 TEAMTLGDRVVVMYGGIAQQIGTPEELYERPANLFVAGFIGSPAMNFFPA
Rv1238|M.tuberculosis_H37Rv TEAMTLGDRVVVMYGGIAQQIGTPEELYERPANLFVAGFIGSPAMNFFPA
MMAR_4203|M.marinum_M TEAMTLGDRVVVMHAGAAQQIGTPQELYERPANLFVAGFIGSPAMNFFPA
MAV_1377|M.avium_104 TEAMTLGDRVVVMHSGVAQQIGTPDELYENPANLFVAGFIGSPAMNFFPA
MLBr_01089|M.leprae_Br4923 TEAMTLGDRVVVMRSGVVQQIGTPDELYERPVNLFVAGFIGSPVMNFFPA
MAB_1375|M.abscessus_ATCC_1997 TEAMTLGDRVVVLRAGKVQQVGAPQELYDTPANLFVAGFIGSPSMNFFPA
Mflv_3854|M.gilvum_PYR-GCK TEAMTLGDRVAVLKKGVLQQVASPRELYDQPVNLFVAGFIGSPPMNFVPA
MUL_2252|M.ulcerans_Agy99 VEAMTMGDRVAVLCDGVLQQCAAPRELYRRPDNVFVAGFIGSPAMNMFIL
.****:****.*: * ** .:* *** * *:********* **:.
Mvan_4484|M.vanbaalenii_PYR-1 ALTEIGVRLPFGEIPLTGQVYERLKGHRTCDDLIAGIRPEHLEDAAVIDA
TH_4034|M.thermoresistible__bu SLTDAGVRLPFGEALLPPDVLDTVRRHEKTDNIIAGLRPEHLEDVSLLDT
MSMEG_5058|M.smegmatis_MC2_155 TRTDVGVRLPFGEVTLTPHMLDLLDKQARPENIIVGIRPEHIEDSALLDG
Mb1270|M.bovis_AF2122/97 RLTAIGLTLPFGEVTLAPEVQGVIAAHPKPENVIVGVRPEHIQDAALIDA
Rv1238|M.tuberculosis_H37Rv RLTAIGLTLPFGEVTLAPEVQGVIAAHPKPENVIVGVRPEHIQDAALIDA
MMAR_4203|M.marinum_M VLTPTGLTLPFGEVTLDPQVQQVIAQHPRPANIIVGIRPEQIQDAALIDA
MAV_1377|M.avium_104 TLTPIGLKVPFGEVMLTPEVQQVIAEHPEPDNVIVGARPEHLSDAALIDG
MLBr_01089|M.leprae_Br4923 KLTPNGLTLPLGELNLSRDVQAMIGQHPVPDRVIVGVRPEHLTDAKLIDA
MAB_1375|M.abscessus_ATCC_1997 TLTDVGVQLPFGEVTLDAGLYAGITARKPSGDVIVGIRPEQFEDAALVDT
Mflv_3854|M.gilvum_PYR-GCK RVTGNEIELPFATVPLRDEWRGSVEEGK---VYIAGIRPGAFEDAEFVDT
MUL_2252|M.ulcerans_Agy99 PIVDSSVLLGDWLIQLPREVTVPA------PEVVVGVRPEHFE-------
. : : * :.* ** :
Mvan_4484|M.vanbaalenii_PYR-1 YARIGALVFDVTVEMVESLGADKYVYFGVDGPGAEAVQLTEVA---AEST
TH_4034|M.thermoresistible__bu YARLGAVTFDVNVELVESLGAEKYVHFRAEGAGAHAAQLAQLA---AESG
MSMEG_5058|M.smegmatis_MC2_155 YARIRALTFSVRADIVESLGADKYVHFTTEGAGAESAQLAELA---ADSG
Mb1270|M.bovis_AF2122/97 YQRIRALTFQVKVNLVESLGADKYLYFTTESPAVHSVQLDELAEVEGESA
Rv1238|M.tuberculosis_H37Rv YQRIRALTFQVKVNLVESLGADKYLYFTTESPAVHSVQLDELAEVEGESA
MMAR_4203|M.marinum_M YQRIRALTFEVNTDLVESLGSDKYVYFSTAGCDVHSAQLDELAGLEGESE
MAV_1377|M.avium_104 YQRIRALTFEVKVDMVESLGADKYVYFSTAAWAAHSTQLDELA---AEAD
MLBr_01089|M.leprae_Br4923 HQHGTVLTFKVKVDLVESLGADKYIYFTTTGCDVYSAQLDELA---SELE
MAB_1375|M.abscessus_ATCC_1997 YKRITGLTLTVNADVVESLGSDKYVHFTTEGGAAHSDELTELA---QESE
Mflv_3854|M.gilvum_PYR-GCK DKASRGVTFDVEIDVTEWLGNEQYAFVPFEATTEISGQLAELAN-ELDSE
MUL_2252|M.ulcerans_Agy99 ---VGNLGVEMEIDVVEELGADAYLYGRISNG----------------GT
: . : ::.* ** : * .
Mvan_4484|M.vanbaalenii_PYR-1 VGENEFVARVPAQSGAAVGRTLQLALNPDNVLLFDPRTGANLS--LPAVG
TH_4034|M.thermoresistible__bu AGENEFIARVSTESKAKKGQSLQLAFNPAKLLVFDADTGANLT--LPPSD
MSMEG_5058|M.smegmatis_MC2_155 AGTNQFIARVSADSRVRTGEQIELAIDTTKLSIFDAATGLNLTRDITPTD
Mb1270|M.bovis_AF2122/97 LHENQFVARVPAESKVAIGQSVELAFDTARLAVFDADSGANLTIPHRA--
Rv1238|M.tuberculosis_H37Rv LHENQFVARVPAESKVAIGQSVELAFDTARLAVFDADSGANLTIPHRA--
MMAR_4203|M.marinum_M VIENRFVARVSAESRATLGRPIELAFDTTRLTVFDADTGANLTNPIAAVQ
MAV_1377|M.avium_104 AHENQFVARVPAESKAAIGQTVELALDTTKLMVFDADSGVNLTVAPSDSP
MLBr_01089|M.leprae_Br4923 VRENQFVARVSAESKMAIGESIELAFGTAKIAVFDADSGVNLTVSAPTDA
MAB_1375|M.abscessus_ATCC_1997 VAENEFVARLSAASKVAEGQPIELIIDTGKLVIFDAESGENLSLAATAE-
Mflv_3854|M.gilvum_PYR-GCK QLRTQICVELDPLSRVRSGDKATLWLDSERLHLFDPHSGDNLTRVSHERP
MUL_2252|M.ulcerans_Agy99 MIDQSVVARADGSNPPERGSRVRLYPQPAQLHFFTVD-GRRIA-------
. .. . * * . .: .* * .::
Mvan_4484|M.vanbaalenii_PYR-1 A----------------
TH_4034|M.thermoresistible__bu A----------------
MSMEG_5058|M.smegmatis_MC2_155 PTEAAGPDAG-------
Mb1270|M.bovis_AF2122/97 -----------------
Rv1238|M.tuberculosis_H37Rv -----------------
MMAR_4203|M.marinum_M -----------------
MAV_1377|M.avium_104 -----------------
MLBr_01089|M.leprae_Br4923 -----------------
MAB_1375|M.abscessus_ATCC_1997 -----------------
Mflv_3854|M.gilvum_PYR-GCK REQNPPESRGRHAATAG
MUL_2252|M.ulcerans_Agy99 -----------------