For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEDWAEIRRLYRFENLSPAAIARRLSLSRNTVAKAINAEAPPRYERAPSTTSAWAQVELAVRLLLGQFPT MPATVLAQRVGWTGGHSWFAENVTRIRPEYPPAGPCDRLVHLPGEQVQCDLWFPGQLVPGHGGGLRSFPV LVMVAAYSRFIAAIMIPSRVTGDLLAGMWQLIARDIGGVPRSLLWDNESGIGQRRHLAQGVAGFCGVLGT RLIQARPYDPETKGVVERANGYLDTSFLPGRTFASPADFNAQLQSWLGAYANNRTHAGTRMIPVDALVAD RAAMAALPPVAPVTGTAVTTRLGRDYYVSVGGNAYSINPEVIGRMITVRASLDRITALCGDRVVADHERL WGTAGLVADPEHVAAAAVLREQFHSRPAASAHLEVSVEVADLGAYDAIFGTGEVA
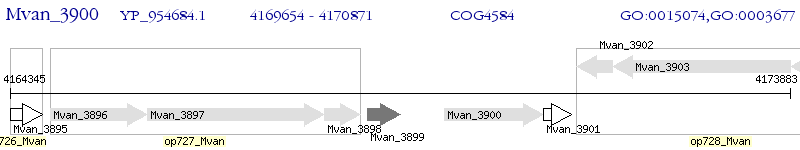
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3900 | - | - | 100% (405) | integrase catalytic subunit |
| M. vanbaalenii PYR-1 | Mvan_0482 | - | 3e-98 | 47.92% (409) | integrase catalytic subunit |
| M. vanbaalenii PYR-1 | Mvan_5931 | - | 3e-08 | 26.34% (391) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2968 | - | 1e-98 | 48.16% (407) | IS1533 transposase |
| M. gilvum PYR-GCK | Mflv_0578 | - | 2e-25 | 46.92% (130) | integrase, catalytic region |
| M. tuberculosis H37Rv | Rv2943 | - | 1e-98 | 48.16% (407) | IS1533 transposase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2619 | - | 6e-05 | 38.18% (55) | transposase for ISMyma06 |
| M. avium 104 | MAV_3533 | - | 6e-08 | 24.00% (350) | transposase |
| M. smegmatis MC2 155 | MSMEG_2151 | - | 3e-09 | 24.61% (386) | transposase IstA protein, putative |
| M. thermoresistible (build 8) | TH_4229 | - | 6e-12 | 26.16% (279) | PUTATIVE putative acyl-CoA synthase |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2968|M.bovis_AF2122/97 --------MLTVEDWAEIRRLHRAEGLPIKMIARVLGISKNTVKSALESN
Rv2943|M.tuberculosis_H37Rv --------MLTVEDWAEIRRLHRAEGLPIKMIARVLGISKNTVKSALESN
Mflv_0578|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3900|M.vanbaalenii_PYR-1 -----------MEDWAEIRRLYRFENLSPAAIARRLSLSRNTVAKAINAE
MAV_3533|M.avium_104 -----------MNIISAYHQVGSYRGAAELCGTTHKTVKR-VIERAEAGG
MSMEG_2151|M.smegmatis_MC2_155 --------MLTWEDDVEVHALR-KRGWTISAIARHTGRDRKTIADYLSGK
TH_4229|M.thermoresistible__bu MAFREVNVNEIREVLRLWLGVAGLPAPGLRTIAAHCGLDRKTVRRYVQAA
MMAR_2619|M.marinum_M -----------MSHANASLTPRGRLRLARCVVDDRWTYARAAERFQCSTA
Mb2968|M.bovis_AF2122/97 QQPKYERAP-QGSIVDAVEPRIRELLQA--YPTMPATVIAERIGWERSIR
Rv2943|M.tuberculosis_H37Rv QQPKYERAP-QGSIVDAVEPRIRELLQA--YPTMPATVIAERIGWERSIR
Mflv_0578|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3900|M.vanbaalenii_PYR-1 APPRYERAPSTTSAWAQVELAVRLLLGQ--FPTMPATVLAQRVGWTGGHS
MAV_3533|M.avium_104 APPREPRPRNLDAFTDLVATRVEKSHGK--MSAKRMLPIARAAGYQGSAR
MSMEG_2151|M.smegmatis_MC2_155 RTPGVRARPGPDPFEPFVDYVAARLTEDPHLWARTLLDELEQLGFPLSYQ
TH_4229|M.thermoresistible__bu HAAGLRRDDDASVIDDGLIGAVAEAVRP-VRPHGHGAAWEQLLAFEGQIR
MMAR_2619|M.marinum_M TAKKWADRYRVGGPEAMVDRSSRPHRSP---GRLPQRRERRIIKIRFTRR
Mb2968|M.bovis_AF2122/97 VLSARVAELRP---VYLPPDPASRTTYV-----------------AGEIA
Rv2943|M.tuberculosis_H37Rv VLSARVAELRP---VYLPPDPASRTTYV-----------------AGEIA
Mflv_0578|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3900|M.vanbaalenii_PYR-1 WFAENVTRIRP---EYPPAGPCDRLVHL-----------------PGEQV
MAV_3533|M.avium_104 NFRRLVAEQKV---LWRNANRHQR--------------------------
MSMEG_2151|M.smegmatis_MC2_155 SLTRNIRARNLRPDCQACRTATERPNSV-----------------IEHAP
TH_4229|M.thermoresistible__bu AWVAGDGEQRPLTITKIHTLLARQGCVVPYRTLHRFASERCGFGRKGTTV
MMAR_2619|M.marinum_M WGPHRIAAHLR---------------------------------------
Mb2968|M.bovis_AF2122/97 QCDFWFPPIELPVGF--------GQTRTAKQLPVLTMVCAYSRWLLAMLL
Rv2943|M.tuberculosis_H37Rv QCDFWFPPIELPVGF--------GQTRTAKQLPVLTMVCAYSRWLLAMLL
Mflv_0578|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3900|M.vanbaalenii_PYR-1 QCDLWFPGQLVP-----------GHGGGLRSFPVLVMVAAYSRFIAAIMI
MAV_3533|M.avium_104 RPAVWSPGDYLVMDW--------AEAAPG--LMVFCAVLAYSRWRFVRFA
MSMEG_2151|M.smegmatis_MC2_155 GDETQWDWLELPDPP--------ESWGWGKTAHLLVGSLAHSGKWRAALT
TH_4229|M.thermoresistible__bu RVADGDPGVECQIDFGYLGMLTDATDGRRRKVHALIFTAVYSRHMFVWLT
MMAR_2619|M.marinum_M -------------------------------LARSTVEAVLRRYRMPLLR
Mb2968|M.bovis_AF2122/97 PSRCAEDLFAGWWRLIE-ALGAVPRVLVWDGEGAIGRWRGGRSELTTECQ
Rv2943|M.tuberculosis_H37Rv PSRCAEDLFAGWWRLIE-ALGAVPRVLVWDGEGAIGRWRGGRSELTTECQ
Mflv_0578|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3900|M.vanbaalenii_PYR-1 PSRVTGDLLAGMWQLIARDIGGVPRSLLWDNESGIG----QRRHLAQGVA
MAV_3533|M.avium_104 ADEKASTTLAMIAEALEAIGGVPAKILADRMGCLKGGVVANVVVPTPDYV
MSMEG_2151|M.smegmatis_MC2_155 PSEDQPQLVAALDRVTRELGGCTR---VWRFDRMATVCDPGSGRVTASFA
TH_4229|M.thermoresistible__bu YSQTLAAVIAGCQAAWAFFGGVFAVLIPDNLKPVIASADALNPQFTRGWL
MMAR_2619|M.marinum_M DLDQASGLPVRRPQPRRYEHPAPGDLVHLDVKKLGRIPDGGGHRKLGRHA
Mb2968|M.bovis_AF2122/97 AFRGTLAAKVLICRPADPEAKGLIERAHDYLERSFLPGRVFASPAD----
Rv2943|M.tuberculosis_H37Rv AFRGTLAAKVLICRPADPEAKGLIERAHDYLERSFLPGRVFASPAD----
Mflv_0578|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3900|M.vanbaalenii_PYR-1 GFCGVLGTRLIQARPYDPETKGVVERANGYLDTSFLPGRTFASPAD----
MAV_3533|M.avium_104 RFASHYGFVPDFCHGADPQSKGIVENLCGYAQDDLAVPLLTEAALAGEQV
MSMEG_2151|M.smegmatis_MC2_155 GVAKHYGVSVAICPPRRGNRKGVVEKVNHTAAQRWWRTLDDELTAEA---
TH_4229|M.thermoresistible__bu DYAGHGGFLTDPARVRSPKDKPRVERVVQYVRRNFWDGETFSSLDEA---
MMAR_2619|M.marinum_M GRRNRSGVGYAFVHHAVDDHSRLAYSEILTDERKDTAAGFWLRANG----
Mb2968|M.bovis_AF2122/97 ----FNAQLGAWL-ALVNTRTRRALGCAPTDRIGADRAAMLSLPPVAPAT
Rv2943|M.tuberculosis_H37Rv ----FNAQLGAWL-ALVNTRTRRALGCAPTDRIGADRAAMLSLPPVAPAT
Mflv_0578|M.gilvum_PYR-GCK -----------------------MLGCAPTDRIGADLAAMLALPPVPPQV
Mvan_3900|M.vanbaalenii_PYR-1 ----FNAQLQSWLGAYANNRTHAGTRMIPVDALVADRAAMAALPPVAPVT
MAV_3533|M.avium_104 DLRALNAQAQLWC-AEVNATVHSEICAVPNDRLVDERTVLRELPSLRPTI
MSMEG_2151|M.smegmatis_MC2_155 ----AQASVDRFARVRGGTRMRATADGRSAVAVVAKTEPLAPVP-ATPYP
TH_4229|M.thermoresistible__bu -----QHVATVWCERTAGTRIHGTTCARPLEVFTAEEQPLLLAMPGVYDV
MMAR_2619|M.marinum_M ----FFVDHGITVKRVLTDNGNCYRSKVFADSLGEEIAHKRTRPYRPQTN
. .
Mb2968|M.bovis_AF2122/97 G-WCTSLRLPRDHYVRCDSNDYSVHPGVIGHRVLVRADLERVHVFC--DG
Rv2943|M.tuberculosis_H37Rv G-WCTSLRLPRDHYVRCDSNDYSVHPGVIGHRVLVRADLERVHVFC--DG
Mflv_0578|M.gilvum_PYR-GCK G-WRNSTRLARDHYVRLDSNDYSVHPGVIGRRIEVVADLDRVQVFC--EG
Mvan_3900|M.vanbaalenii_PYR-1 G-TAVTTRLGRDYYVSVGGNAYSINPEVIGRMITVRASLDRITALC--GD
MAV_3533|M.avium_104 GSGSVRRKVDGLSCIRYGSARYSVPQRLVGATVAVVVDHGALILLEPATG
MSMEG_2151|M.smegmatis_MC2_155 VIISQTRTASRQALVSYRGNRYSVPPELAAAQVVVSHPVGAQFCDIATVA
TH_4229|M.thermoresistible__bu P-VFKAVKVHRDFHAEVAKALYSLPECWIGTTLDVRADGELVKFYH--RG
MMAR_2619|M.marinum_M G--------KVERFNRTLASEWAYAQTYLSEVQRAATYQDWLHYYN----
:: . .
Mb2968|M.bovis_AF2122/97 ELVADHERIWAV--HQTVSDPAHVEAAKVLRR------------RHFSAA
Rv2943|M.tuberculosis_H37Rv ELVADHERIWAV--HQTVSDPAHVEAAKVLRR------------RHFSAA
Mflv_0578|M.gilvum_PYR-GCK KTVAEHERVWAW--HQTITDPEHREAANMLRR------------NRISAL
Mvan_3900|M.vanbaalenii_PYR-1 RVVADHERLWGT--AGLVADPEHVAAAAVLRE------------QFHS--
MAV_3533|M.avium_104 VIVAEHELVSPG--EVSILDEHYDGPRPAPSRGPRPK--TQAE-KRFCAL
MSMEG_2151|M.smegmatis_MC2_155 GIVIARHRMAADGLGVMVRDSGHVIALDTMAMATASTGRPHRR-KERIPP
TH_4229|M.thermoresistible__bu TLVKVHPRQPAGGRSTDRGDLPEHKAGYALRDLAALIAACAAHGPNIGIY
MMAR_2619|M.marinum_M -----HHRPHTGIGGKTPIERLRVHNLPVKNT------------------
: :
Mb2968|M.bovis_AF2122/97 SPVVEP----QVQVRS---LSDYDDALGVDIDGG--VA------------
Rv2943|M.tuberculosis_H37Rv SPVVEP----QVQVRS---LSDYDDALGVDIDGG--VA------------
Mflv_0578|M.gilvum_PYR-GCK RPVTDSDQQVLVEQRR---LSDYDAALGIDLGEGGLVS------------
Mvan_3900|M.vanbaalenii_PYR-1 RPAASAHLEVSVEVAD---LGAYDAIFGTGEVA-----------------
MAV_3533|M.avium_104 GTEAQQFLVGAAAIGNTRLKSELDILLGLGAAHGEQALIDALRRAVAFRR
MSMEG_2151|M.smegmatis_MC2_155 GPAAKAAAAQLLHLNDANQEAAESSTWSTDSTVIDLSAYERAAQKRTIQ-
TH_4229|M.thermoresistible__bu AERILDDPLPWTRMRTVYRLLGLVRRYGAQRVEAACSLSLDLDVVSVTKI
MMAR_2619|M.marinum_M --------------------------------------------------
Mb2968|M.bovis_AF2122/97 --------------------------------------------------
Rv2943|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0578|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3900|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3533|M.avium_104 FRAADVRSILAAGAGTPQPRPAGDALVLDLPTVETRSLEAYKINTTDGTA
MSMEG_2151|M.smegmatis_MC2_155 --------------------------------------------------
TH_4229|M.thermoresistible__bu ASMLERATETCAPALPRAVGQEATRFSRDPSEFSSTPTPLTVISHSESEE
MMAR_2619|M.marinum_M --------------------------------------------------
Mb2968|M.bovis_AF2122/97 --
Rv2943|M.tuberculosis_H37Rv --
Mflv_0578|M.gilvum_PYR-GCK --
Mvan_3900|M.vanbaalenii_PYR-1 --
MAV_3533|M.avium_104 S-
MSMEG_2151|M.smegmatis_MC2_155 --
TH_4229|M.thermoresistible__bu TR
MMAR_2619|M.marinum_M --