For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSPWVVAAVTVALISLSAFFVAVEFALIAARRSRLEDAAATSAAARAALRSAGELSLLLAGAQLGITVC TLLLGAITKPAVHHAITPLIRDLGAPAWAADVGGFVLALLIVTFLHLVVGEMAPKSWAIAHPERSATLLA LPMRGFMFVTRPLLRGLNAAANWCLRRVGVTPVDELADTQDPDTLRQLVEHSATVGTLDERYHANLTSAL ELEALTVGQLARRDMRYSEVPADATAAQIQATAKATGHLRLLVRDGTATVGVLHVRDSLSNTGAARDLMR PILAMPENTPVYEALRTMRETRNHLVVVDGPSGRGLLTLTDLLYRLLPTEAA
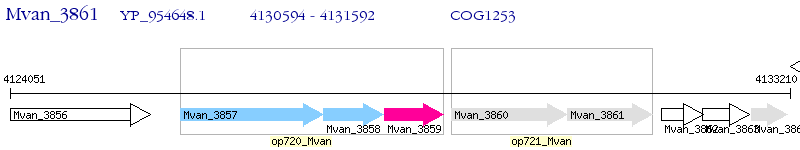
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3861 | - | - | 100% (332) | hypothetical protein Mvan_3861 |
| M. vanbaalenii PYR-1 | Mvan_3095 | - | 1e-47 | 37.39% (337) | hypothetical protein Mvan_3095 |
| M. vanbaalenii PYR-1 | Mvan_3094 | - | 3e-33 | 31.99% (322) | hypothetical protein Mvan_3094 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1872c | - | 1e-44 | 36.47% (340) | hypothetical protein Mb1872c |
| M. gilvum PYR-GCK | Mflv_2682 | - | 1e-162 | 85.84% (332) | hypothetical protein Mflv_2682 |
| M. tuberculosis H37Rv | Rv1841c | - | 2e-43 | 36.47% (340) | hypothetical protein Rv1841c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2410c | - | 8e-48 | 37.61% (343) | hypothetical protein MAB_2410c |
| M. marinum M | MMAR_2715 | - | 5e-42 | 34.90% (341) | hypothetical protein MMAR_2715 |
| M. avium 104 | MAV_2879 | - | 4e-42 | 34.55% (330) | CBS domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_3638 | - | 3e-46 | 37.26% (314) | CBS domain-containing protein |
| M. thermoresistible (build 8) | TH_0286 | - | 2e-48 | 38.62% (334) | CBS domain protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1872c|M.bovis_AF2122/97 MD-VLSAVLLALLLIGANAFFVGAEFALISARRDRLEALAEQGKATAVTV
Rv1841c|M.tuberculosis_H37Rv MD-VLSAVLLALLLIGANAFFVGAEFALISARRDRLEALAEQGKATAVTV
MMAR_2715|M.marinum_M MN-PVLAVALAVLLIAANAFFVGAEFALISARRDRLEALAEQGRTSAVTV
MAV_2879|M.avium_104 MS-DTMGVLLACLLIGVNAFFVGAEFSLISARRDRLEALAEQGRAAAVTV
MSMEG_3638|M.smegmatis_MC2_155 MG-DVFGVLLTVLLLAANAFFVAAEFALISARRDRLQALAEQGKKSAITV
TH_0286|M.thermoresistible__bu MG-DLFGVLLTVLLLGANAFFVAAEFSLISARRDRLEALAEQGKKSAVTV
MAB_2410c|M.abscessus_ATCC_199 MGSDILGVLLTAALLLANAFFVGAEFALISARRDRLEALAEAGKKRAVTV
Mvan_3861|M.vanbaalenii_PYR-1 MSSPWVVAAVTVALISLSAFFVAVEFALIAARRSRLEDAAATS-AAARAA
Mflv_2682|M.gilvum_PYR-GCK MSSPWVIAGITAGLIGLSAFFVAVEFALIAARRNRLEDAAATS-AAARAA
*. . :: *: .****..**:**:***.**: * . * :.
Mb1872c|M.bovis_AF2122/97 IRAGEQLPAMLTGAQLGVTVSSILLGRVGEPAVVKLLQLSFGLSGVPPAL
Rv1841c|M.tuberculosis_H37Rv IRAGEQLPAMLTGAQLGVTVSSILLGRVGEPAVVKLLQLSFGLSGVPPAL
MMAR_2715|M.marinum_M IRAAEQLPSMLAGAQFGVTVSSILLGRIAEPAAAQTLQRWFGLAGLDPRL
MAV_2879|M.avium_104 IRASEQLPSMLAGAQLGVTAASLLLGRIGESAVSNLLRTVLGLTRIHPAL
MSMEG_3638|M.smegmatis_MC2_155 IRAGEHLSLMLAGSQLGITICSILLGRVGEPAVAHLLEKPFALIGVSDTV
TH_0286|M.thermoresistible__bu IRAGEQLSLMLAGSQLGITICSILLGRIGEPAVAHLLEKPFDLLGVPDAV
MAB_2410c|M.abscessus_ATCC_199 MMAGQNLSLMLAGAQLGITICSILLGRVGEPAVAHLLERPFELLGAPAAV
Mvan_3861|M.vanbaalenii_PYR-1 LRSAGELSLLLAGAQLGITVCTLLLGAITKPAVHHAITPLIRDLGAPAWA
Mflv_2682|M.gilvum_PYR-GCK LRSAGELSLLLAGAQLGITACTLLLGAITKPAVHHAITPAIGDLGAPGWV
: :. .*. :*:*:*:*:* .::*** : :.*. : : :
Mb1872c|M.bovis_AF2122/97 LHTLSLAVALAIVVALHVLLGEMVPKNIALAGPERTAMLLVPPYLVYVRL
Rv1841c|M.tuberculosis_H37Rv LHTLS----LAIVVALHVLLGEMVPKNIALAGPERTAMLLVPPYLVYVRL
MMAR_2715|M.marinum_M LHTLSFVVALAIVVTLHVLLGEMVPKNIALAGPERTAMLLVPAYLPYVRA
MAV_2879|M.avium_104 LHSLSLAIALAIVVTLHVLLGEMVPKNIALAGPERTAMLLVPPYLAYVRA
MSMEG_3638|M.smegmatis_MC2_155 LHTVSFVVALAVVVILHVLLGEMVPKNIAIAGPETAAMLLIPPYLLYIRA
TH_0286|M.thermoresistible__bu LHSVSFVVALSIVVILHVLLGEMVPKNIAIAGPEKSAMLLVPTFLVYMRA
MAB_2410c|M.abscessus_ATCC_199 THTVAFVISLSIVVLLHVLLGEMVPKNIAIAGPESAAMLLIPPYLVYIRL
Mvan_3861|M.vanbaalenii_PYR-1 ADVGGFVLALLIVTFLHLVVGEMAPKSWAIAHPERSATLLALPMRGFMFV
Mflv_2682|M.gilvum_PYR-GCK ADVGGFVLALLIVTFLHLVIGEMAPKSWAIAHPERSATLLALPMRGFMFL
. . * :*. **:::***.**. *:* ** :* ** . ::
Mb1872c|M.bovis_AF2122/97 ARPFIAFYNNCANAILRLVGVQPKDELDIAVSTAELSEMIAESLSEGLLD
Rv1841c|M.tuberculosis_H37Rv ARPFIAFYNNCANAILRLVGVQPKDELDIAVSTAELSEMIAESLSEGLLD
MMAR_2715|M.marinum_M ARPIIFLYNKCANAILRIFGVQPKDELDITVSTVELSEMIAESVSEGLLD
MAV_2879|M.avium_104 ARPFIAFYNRCASVMVRALGVEPKEELEITVSPVELSEMIAESESEGLLD
MSMEG_3638|M.smegmatis_MC2_155 ARPFIAFYNWCANATLRAFGVEPRDELASAVSTVELSEMIAESVSEGLLD
TH_0286|M.thermoresistible__bu VRPLIAFYNWCANATLRAFRVQPRDELDVTVSTVELSEMIAESVSEGLLD
MAB_2410c|M.abscessus_ATCC_199 ARPLIAFYNWCANTALRLVKVPVKDELEVTVSTVELSEMIAESLSEGLLD
Mvan_3861|M.vanbaalenii_PYR-1 TRPLLRGLNAAANWCLRRVGVTPVDELADTQDPDTLRQLVEHSATVGTLD
Mflv_2682|M.gilvum_PYR-GCK TRPMLRALNRAANWCLRRVGVTPVDELADSQDPDALRQLVEHSATVGTLD
.**:: * .*. :* . * :** : .. * ::: .* : * **
Mb1872c|M.bovis_AF2122/97 HEEHTRLTRALRIRTRLVADVAVPLVNIRAVQVSAVGSGPTIGGVEQALA
Rv1841c|M.tuberculosis_H37Rv HEEHTRLTRALRIRTRLVADVAVPLVNIRAVQVSAVGSGPTIGGVEQALA
MMAR_2715|M.marinum_M PEEHTRLTRALRIRTRVVDDVAVPLPNIRAVPVAAAGSGPTIGAVEQALA
MAV_2879|M.avium_104 REEHTRLTRALQLRHRVVGDVAVPLARVHAVPVAAPGSGPTVGAVEQALA
MSMEG_3638|M.smegmatis_MC2_155 TEEHSRLTRALQTRTRTVTDVAMPLSGIHAVSVAGPGCGPTVGAVEQALA
TH_0286|M.thermoresistible__bu PEEHSRLTRALQIRTRVVNDVAMPLDQIRTVPVAGPGRGPTVGAVEQALK
MAB_2410c|M.abscessus_ATCC_199 EEEHTRLRRALQIQHRVVADVVVPANQIRSIAVSGSAQAPSVTAVERAVT
Mvan_3861|M.vanbaalenii_PYR-1 ERYHANLTSALELEALTVGQLARRDMRYSEVPADA-----TAAQIQATAK
Mflv_2682|M.gilvum_PYR-GCK ERYHANLASALELEALTVGQVARHDARYSDVPVDA-----TPAQIQETAR
. *:.* **. . * ::. : . . : :: :
Mb1872c|M.bovis_AF2122/97 QTGYSRFPVVDRGGRFIGYLHIKDVLTLGDNPQTVIDLAVVRPLPRVPQS
Rv1841c|M.tuberculosis_H37Rv QTGYSRFPVVDRGGRFIGYLHIKDVLTLGDNPQTVIDLAVVRPLPRVPQS
MMAR_2715|M.marinum_M ETGYSRFPVLGLDQHFIGYLHIKDVLALGDDPATVIDLALVRPLPQLAKS
MAV_2879|M.avium_104 QTGYSRFPVTNPTGDFIGYLHIKDVLTLDDDPATVINLAKVRPLPRLPRS
MSMEG_3638|M.smegmatis_MC2_155 ETGYSRFPVTDASGTFIGYLHIKDVLLQVNEPDSVVDLAMVRPLPQVPAS
TH_0286|M.thermoresistible__bu ATGYSRFPVTAPSGEFLGYLHIKDVLPFADDPETVLDLSMVRPLPALPAS
MAB_2410c|M.abscessus_ATCC_199 ETGYSRFPVIGPRGDYLGYLHIKDVLTMGDDPEAIVDPAFIRPLPRLHGS
Mvan_3861|M.vanbaalenii_PYR-1 ATGHLRLLVRD-GTATVGVLHVRDSLSNTGAAR-----DLMRPILAMPEN
Mflv_2682|M.gilvum_PYR-GCK ATGHLRLLVRD-GGATVGVVHVRDCLTGDATAR-----ELMRPVLTMPDS
**: *: * :* :*::* * . :**: : .
Mb1872c|M.bovis_AF2122/97 LPLADALSRMRRINSHLALVTAD--------------NGSVVGMVALEDV
Rv1841c|M.tuberculosis_H37Rv LPLADALSRMRRINSHLALVTAD--------------NGSVVGMVALEDV
MMAR_2715|M.marinum_M LPLPDALSRMRRSNSHLALVTSD--------------DGAVAGMVALEDL
MAV_2879|M.avium_104 LPLADGLSRMRRSNSHLALVTDQ--------------GGAVVAMVAMEDL
MSMEG_3638|M.smegmatis_MC2_155 LPVPDALSRMRRSNSHLALVVDDSLIDVHGSTAHTAADNTVTAMVALEDL
TH_0286|M.thermoresistible__bu LPLPEALSRMRRNNSHLALVTTD---------------GRVTAMVALEDL
MAB_2410c|M.abscessus_ATCC_199 VPLPEALSTLRRSNSHLALMSTT--------------DGSVVGLVTLEDL
Mvan_3861|M.vanbaalenii_PYR-1 TPVYEALRTMRETRNHLVVVDGP----------------SGRGLLTLTDL
Mflv_2682|M.gilvum_PYR-GCK TPVYEALQLMRQTRTHLVVVDGP----------------SGPGLLTLTDL
*: :.* :*. ..**.:: .:::: *:
Mb1872c|M.bovis_AF2122/97 VEDLVGTMRDGTHR-
Rv1841c|M.tuberculosis_H37Rv VEDLVGTMRDGTHR-
MMAR_2715|M.marinum_M VEDLVGTLSDGKHRN
MAV_2879|M.avium_104 VEDLVGTMREA----
MSMEG_3638|M.smegmatis_MC2_155 VEDLVGTVRDGTHRV
TH_0286|M.thermoresistible__bu VEDLVGTVRDGTHRV
MAB_2410c|M.abscessus_ATCC_199 VEDLVGTVRDGTHRI
Mvan_3861|M.vanbaalenii_PYR-1 LYRLLPTEAA-----
Mflv_2682|M.gilvum_PYR-GCK MYRLLPTEAA-----
: *: *