For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGDVFGVLLTVLLLAANAFFVAAEFALISARRDRLQALAEQGKKSAITVIRAGEHLSLMLAGSQLGITIC SILLGRVGEPAVAHLLEKPFALIGVSDTVLHTVSFVVALAVVVILHVLLGEMVPKNIAIAGPETAAMLLI PPYLLYIRAARPFIAFYNWCANATLRAFGVEPRDELASAVSTVELSEMIAESVSEGLLDTEEHSRLTRAL QTRTRTVTDVAMPLSGIHAVSVAGPGCGPTVGAVEQALAETGYSRFPVTDASGTFIGYLHIKDVLLQVNE PDSVVDLAMVRPLPQVPASLPVPDALSRMRRSNSHLALVVDDSLIDVHGSTAHTAADNTVTAMVALEDLV EDLVGTVRDGTHRV
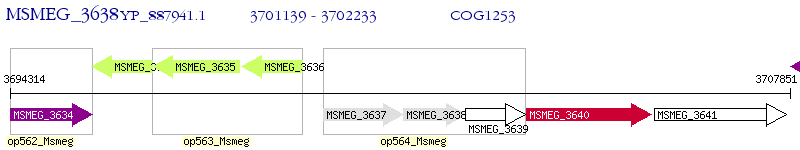
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3638 | - | - | 100% (364) | CBS domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_3637 | - | 6e-42 | 32.40% (358) | CBS domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_4495 | - | 4e-11 | 25.00% (184) | CBS domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1872c | - | 1e-134 | 69.75% (357) | hypothetical protein Mb1872c |
| M. gilvum PYR-GCK | Mflv_3365 | - | 1e-150 | 75.20% (367) | hypothetical protein Mflv_3365 |
| M. tuberculosis H37Rv | Rv1841c | - | 1e-131 | 69.19% (357) | hypothetical protein Rv1841c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2410c | - | 1e-132 | 65.75% (362) | hypothetical protein MAB_2410c |
| M. marinum M | MMAR_2715 | - | 1e-132 | 68.60% (363) | hypothetical protein MMAR_2715 |
| M. avium 104 | MAV_2879 | - | 1e-128 | 65.74% (359) | CBS domain-containing protein |
| M. thermoresistible (build 8) | TH_0286 | - | 1e-156 | 78.85% (364) | CBS domain protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3095 | - | 1e-151 | 75.75% (367) | hypothetical protein Mvan_3095 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3638|M.smegmatis_MC2_155 MG-DVFGVLLTVLLLAANAFFVAAEFALISARRDRLQALAEQGKKSAITV
TH_0286|M.thermoresistible__bu MG-DLFGVLLTVLLLGANAFFVAAEFSLISARRDRLEALAEQGKKSAVTV
Mflv_3365|M.gilvum_PYR-GCK MG-DIFGVLLTFVLLGANAFFVASEFALISARRDRLEALAEQGKSSAVTV
Mvan_3095|M.vanbaalenii_PYR-1 MG-DVFGVLLTFVLLGANAFFVASEFALISARRDRLEALAEQGKSSAVTV
MAB_2410c|M.abscessus_ATCC_199 MGSDILGVLLTAALLLANAFFVGAEFALISARRDRLEALAEAGKKRAVTV
Mb1872c|M.bovis_AF2122/97 MD-VLSAVLLALLLIGANAFFVGAEFALISARRDRLEALAEQGKATAVTV
Rv1841c|M.tuberculosis_H37Rv MD-VLSAVLLALLLIGANAFFVGAEFALISARRDRLEALAEQGKATAVTV
MMAR_2715|M.marinum_M MN-PVLAVALAVLLIAANAFFVGAEFALISARRDRLEALAEQGRTSAVTV
MAV_2879|M.avium_104 MS-DTMGVLLACLLIGVNAFFVGAEFSLISARRDRLEALAEQGRAAAVTV
*. .* *: *: .*****.:**:*********:**** *: *:**
MSMEG_3638|M.smegmatis_MC2_155 IRAGEHLSLMLAGSQLGITICSILLGRVGEPAVAHLLEKPFALIGVSDTV
TH_0286|M.thermoresistible__bu IRAGEQLSLMLAGSQLGITICSILLGRIGEPAVAHLLEKPFDLLGVPDAV
Mflv_3365|M.gilvum_PYR-GCK IRAGENLSLMLAGSQLGITICSILLGRVAEPAVAHLLEKPFDLVGIPDAL
Mvan_3095|M.vanbaalenii_PYR-1 IRAGENLSLMLAGSQLGITVCSILLGRVAEPAVAHLLEKPFGLVGIPDAV
MAB_2410c|M.abscessus_ATCC_199 MMAGQNLSLMLAGAQLGITICSILLGRVGEPAVAHLLERPFELLGAPAAV
Mb1872c|M.bovis_AF2122/97 IRAGEQLPAMLTGAQLGVTVSSILLGRVGEPAVVKLLQLSFGLSGVPPAL
Rv1841c|M.tuberculosis_H37Rv IRAGEQLPAMLTGAQLGVTVSSILLGRVGEPAVVKLLQLSFGLSGVPPAL
MMAR_2715|M.marinum_M IRAAEQLPSMLAGAQFGVTVSSILLGRIAEPAAAQTLQRWFGLAGLDPRL
MAV_2879|M.avium_104 IRASEQLPSMLAGAQLGVTAASLLLGRIGESAVSNLLRTVLGLTRIHPAL
: *.::*. **:*:*:*:* .*:****:.*.*. : *. : * :
MSMEG_3638|M.smegmatis_MC2_155 LHTVSFVVALAVVVILHVLLGEMVPKNIAIAGPETAAMLLIPPYLLYIRA
TH_0286|M.thermoresistible__bu LHSVSFVVALSIVVILHVLLGEMVPKNIAIAGPEKSAMLLVPTFLVYMRA
Mflv_3365|M.gilvum_PYR-GCK LHTVSFLVALSIVVTLHVLLGEMVPKNIAIAGPESTAMLLIPVYLLYIRI
Mvan_3095|M.vanbaalenii_PYR-1 LHTVSFLVALSIVVTLHVLLGEMVPKNIAIAGPESTAMLLIPVYLVYIRM
MAB_2410c|M.abscessus_ATCC_199 THTVAFVISLSIVVLLHVLLGEMVPKNIAIAGPESAAMLLIPPYLVYIRL
Mb1872c|M.bovis_AF2122/97 LHTLSLAVALAIVVALHVLLGEMVPKNIALAGPERTAMLLVPPYLVYVRL
Rv1841c|M.tuberculosis_H37Rv LHTLS----LAIVVALHVLLGEMVPKNIALAGPERTAMLLVPPYLVYVRL
MMAR_2715|M.marinum_M LHTLSFVVALAIVVTLHVLLGEMVPKNIALAGPERTAMLLVPAYLPYVRA
MAV_2879|M.avium_104 LHSLSLAIALAIVVTLHVLLGEMVPKNIALAGPERTAMLLVPPYLAYVRA
*::: *::** **************:**** :****:* :* *:*
MSMEG_3638|M.smegmatis_MC2_155 ARPFIAFYNWCANATLRAFGVEPRDELASAVSTVELSEMIAESVSEGLLD
TH_0286|M.thermoresistible__bu VRPLIAFYNWCANATLRAFRVQPRDELDVTVSTVELSEMIAESVSEGLLD
Mflv_3365|M.gilvum_PYR-GCK ARPFIAFYNWCANSTLRAFGVEPKDELDVTVSTVELSEMIAESLSEGLLD
Mvan_3095|M.vanbaalenii_PYR-1 ARPFIAFYNWCANATLRAFGVEPKDELDVTVSTVELSEMIAESLSEGLLD
MAB_2410c|M.abscessus_ATCC_199 ARPLIAFYNWCANTALRLVKVPVKDELEVTVSTVELSEMIAESLSEGLLD
Mb1872c|M.bovis_AF2122/97 ARPFIAFYNNCANAILRLVGVQPKDELDIAVSTAELSEMIAESLSEGLLD
Rv1841c|M.tuberculosis_H37Rv ARPFIAFYNNCANAILRLVGVQPKDELDIAVSTAELSEMIAESLSEGLLD
MMAR_2715|M.marinum_M ARPIIFLYNKCANAILRIFGVQPKDELDITVSTVELSEMIAESVSEGLLD
MAV_2879|M.avium_104 ARPFIAFYNRCASVMVRALGVEPKEELEITVSPVELSEMIAESESEGLLD
.**:* :** **. :* . * ::** :**..********* ******
MSMEG_3638|M.smegmatis_MC2_155 TEEHSRLTRALQTRTRTVTDVAMPLSGIHAVSVAGPGCGPTVGAVEQALA
TH_0286|M.thermoresistible__bu PEEHSRLTRALQIRTRVVNDVAMPLDQIRTVPVAGPGRGPTVGAVEQALK
Mflv_3365|M.gilvum_PYR-GCK PEEHTRLTRALQIRNRVVDDVAMPLHRIRAVPAAHDGAGPTVGALEEALR
Mvan_3095|M.vanbaalenii_PYR-1 PEEHTRLTRALQIRNRVVNDVAMPLHQIRAVPVAQEGAGPTVGALEEALK
MAB_2410c|M.abscessus_ATCC_199 EEEHTRLRRALQIQHRVVADVVVPANQIRSIAVSGSAQAPSVTAVERAVT
Mb1872c|M.bovis_AF2122/97 HEEHTRLTRALRIRTRLVADVAVPLVNIRAVQVSAVGSGPTIGGVEQALA
Rv1841c|M.tuberculosis_H37Rv HEEHTRLTRALRIRTRLVADVAVPLVNIRAVQVSAVGSGPTIGGVEQALA
MMAR_2715|M.marinum_M PEEHTRLTRALRIRTRVVDDVAVPLPNIRAVPVAAAGSGPTIGAVEQALA
MAV_2879|M.avium_104 REEHTRLTRALQLRHRVVGDVAVPLARVHAVPVAAPGSGPTVGAVEQALA
***:** ***: : * * **.:* :::: .: . .*:: .:*.*:
MSMEG_3638|M.smegmatis_MC2_155 ETGYSRFPVTDASGTFIGYLHIKDVLLQVN---EPDSVVDLAMVRPLPQV
TH_0286|M.thermoresistible__bu ATGYSRFPVTAPSGEFLGYLHIKDVLPFAD---DPETVLDLSMVRPLPAL
Mflv_3365|M.gilvum_PYR-GCK ETGYSRFPVADSTGAFVGYLHIKDVLPLVNGNYDSSTAVEASLVRPLPRV
Mvan_3095|M.vanbaalenii_PYR-1 ETGYSRFPVADPTGAYIGYLHIKDVLPLVAGDSGGTAVIETSMVRPLPRV
MAB_2410c|M.abscessus_ATCC_199 ETGYSRFPVIGPRGDYLGYLHIKDVLTMGD---DPEAIVDPAFIRPLPRL
Mb1872c|M.bovis_AF2122/97 QTGYSRFPVVDRGGRFIGYLHIKDVLTLGD---NPQTVIDLAVVRPLPRV
Rv1841c|M.tuberculosis_H37Rv QTGYSRFPVVDRGGRFIGYLHIKDVLTLGD---NPQTVIDLAVVRPLPRV
MMAR_2715|M.marinum_M ETGYSRFPVLGLDQHFIGYLHIKDVLALGD---DPATVIDLALVRPLPQL
MAV_2879|M.avium_104 QTGYSRFPVTNPTGDFIGYLHIKDVLTLDD---DPATVINLAKVRPLPRL
******** ::********* : :: : :**** :
MSMEG_3638|M.smegmatis_MC2_155 PASLPVPDALSRMRRSNSHLALVVDDSLIDVHGSTAHTAADNTVTAMVAL
TH_0286|M.thermoresistible__bu PASLPLPEALSRMRRNNSHLALVTTD---------------GRVTAMVAL
Mflv_3365|M.gilvum_PYR-GCK PASLPLPDALTRLRRTNSHLALVTAA--------------DGDATAVVAL
Mvan_3095|M.vanbaalenii_PYR-1 PASLPLPDALTRLRRTNSHLALVTAG--------------DGTPTAMVAL
MAB_2410c|M.abscessus_ATCC_199 HGSVPLPEALSTLRRSNSHLALMSTT--------------DGSVVGLVTL
Mb1872c|M.bovis_AF2122/97 PQSLPLADALSRMRRINSHLALVTAD--------------NGSVVGMVAL
Rv1841c|M.tuberculosis_H37Rv PQSLPLADALSRMRRINSHLALVTAD--------------NGSVVGMVAL
MMAR_2715|M.marinum_M AKSLPLPDALSRMRRSNSHLALVTSD--------------DGAVAGMVAL
MAV_2879|M.avium_104 PRSLPLADGLSRMRRSNSHLALVTDQ--------------GGAVVAMVAM
*:*:.:.*: :** ******: . ..:*::
MSMEG_3638|M.smegmatis_MC2_155 EDLVEDLVGTVRDGTHRV
TH_0286|M.thermoresistible__bu EDLVEDLVGTVRDGTHRV
Mflv_3365|M.gilvum_PYR-GCK EDLVEDLVGTVRDSTHRV
Mvan_3095|M.vanbaalenii_PYR-1 EDLVEDLVGTVRDSTHRI
MAB_2410c|M.abscessus_ATCC_199 EDLVEDLVGTVRDGTHRI
Mb1872c|M.bovis_AF2122/97 EDVVEDLVGTMRDGTHR-
Rv1841c|M.tuberculosis_H37Rv EDVVEDLVGTMRDGTHR-
MMAR_2715|M.marinum_M EDLVEDLVGTLSDGKHRN
MAV_2879|M.avium_104 EDLVEDLVGTMREA----
**:*******: :.