For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DGVTAIATSGHVLLAVFVSALAGLVSFASPCVVPLVPGYLSYLAAVVGVDDQARAQPLGRARLRLAGAAG LFVAGFTAVFLMGTVAILGLTTTLITNGVLLQRIGGVVTIVMGLVFMGFFPALQRDARFTPTSLSTLWGA PLLGAVFGLGWTPCLGPTLTGVIAVASASEGPNVVRGIVLVVAYCVGLGAPFVLFALASGRAMRALGWLR RNARRIQVFGGVLLVAVGIALVTGLWNEVVSWVRDAFVSNTTLPI*
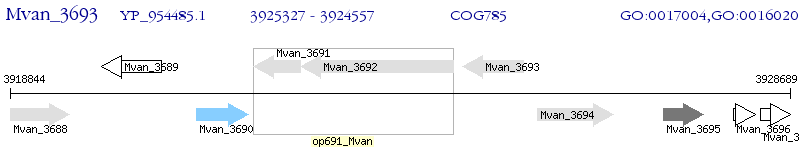
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3693 | - | - | 100% (256) | cytochrome c biogenesis protein, transmembrane region |
| M. vanbaalenii PYR-1 | Mvan_0867 | - | e-102 | 70.23% (262) | cytochrome c biogenesis protein, transmembrane region |
| M. vanbaalenii PYR-1 | Mvan_5557 | - | 1e-12 | 32.05% (234) | cytochrome c biogenesis protein, transmembrane region |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0540 | ccdA | 2e-99 | 70.43% (257) | cytochrome C-type biogenesis protein CcdA |
| M. gilvum PYR-GCK | Mflv_2825 | - | 1e-139 | 97.67% (257) | cytochrome c biogenesis protein, transmembrane region |
| M. tuberculosis H37Rv | Rv0527 | ccdA | 2e-99 | 70.43% (257) | cytochrome C-type biogenesis protein CcdA |
| M. leprae Br4923 | MLBr_02411 | - | 1e-86 | 61.15% (260) | putative cytochrome C-type biogenesis protein |
| M. abscessus ATCC 19977 | MAB_3975c | - | 9e-97 | 69.96% (253) | putative cytochrome C biogenesis protein CcdA |
| M. marinum M | MMAR_0873 | ccsA | 3e-96 | 66.28% (261) | cytochrome C-type biogenesis protein, CcsA |
| M. avium 104 | MAV_4618 | - | 1e-98 | 69.50% (259) | cytochrome C biogenesis protein transmembrane region |
| M. smegmatis MC2 155 | MSMEG_0972 | - | 1e-103 | 73.95% (261) | cytochrome C biogenesis protein transmembrane region |
| M. thermoresistible (build 8) | TH_2250 | ccsA | 1e-101 | 68.63% (271) | POSSIBLE CYTOCHROME C-TYPE BIOGENESIS PROTEIN CCDA |
| M. ulcerans Agy99 | MUL_0626 | ccsA | 1e-95 | 65.90% (261) | cytochrome C-type biogenesis protein, CcsA |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0873|M.marinum_M ------MTGFAEVAAAGPLLVALGLCVLAGLVSFASPCVVPLVPGYLSYL
MUL_0626|M.ulcerans_Agy99 ------MTGFAEVAAAGPLLVALGLCVLAGLVSFASPCVVPLVPGYLSYL
MAV_4618|M.avium_104 ------MTGLTQIAAAGPLLAALGVCLLAGLVSFASPCVVPLVPGYLSYL
Mb0540|M.bovis_AF2122/97 ------MTGFTEIAAVGPLLVAVGVCLLAGLVSFASPCVVPLVPGYLSYL
Rv0527|M.tuberculosis_H37Rv ------MTGFTEIAAVGPLLVAVGVCLLAGLVSFASPCVVPLVPGYLSYL
MLBr_02411|M.leprae_Br4923 ------MTGFTEIAAAGPLLVALGVCMLAGLVSFVSPCVVPLVPGYLSYL
MSMEG_0972|M.smegmatis_MC2_155 ------MTGFAEIAAAGPVLLALGVSVLAGLVSFASPCVVPLVPGYLSYL
TH_2250|M.thermoresistible__bu ------MTGFAELAAAGPVLVAVAISILAGLVSFASPCVVPLVPGYLSYL
MAB_3975c|M.abscessus_ATCC_199 MILADAGSAFQDAATSGPLLLALGACALAGLVSFASPCVIPLVPGYLSYL
Mvan_3693|M.vanbaalenii_PYR-1 ------MDGVTAIATSGHVLLAVFVSALAGLVSFASPCVVPLVPGYLSYL
Mflv_2825|M.gilvum_PYR-GCK ------MDGVTAIATSGHVLLAVFVSALAGLVSFASPCVVPLVPGYLSYL
.. *: * :* *: . *******.****:**********
MMAR_0873|M.marinum_M AAVVGVDGSGDGRAAGVGVKAPA----------GVRWRVAGSALLFVAGF
MUL_0626|M.ulcerans_Agy99 AAVVGVDGSGDGRAAGVGVKAPA----------GVRWRVAGSALLFVAGF
MAV_4618|M.avium_104 AALVGVE---EQPQAGA-VQAPP----------GARWRVAGSAALFVAGF
Mb0540|M.bovis_AF2122/97 AAVVGV---DEQLP-AGVVKPPV----------AARWRVAGSAALFVAGF
Rv0527|M.tuberculosis_H37Rv AAVVGV---DEQLP-AGVVKPPV----------AARWRVAGSAALFVAGF
MLBr_02411|M.leprae_Br4923 AAVVGVS-HEETQPGAGVIKTPP----------AARWRVAGSAVLFVAGF
MSMEG_0972|M.smegmatis_MC2_155 AAVVGVDDD--PVRTGGER----------VALRSARLRVAGAAALFVAGF
TH_2250|M.thermoresistible__bu AAVVGVPKEAAPVVTGGPRRDPPADSGAGMSPRSARLRVTGAAALFVAGF
MAB_3975c|M.abscessus_ATCC_199 AAVVGAD--NGPENGAGAQPH--------------RLRVTGSALLFVAGF
Mvan_3693|M.vanbaalenii_PYR-1 AAVVGVDD-QARAQPLG----------------RARLRLAGAAGLFVAGF
Mflv_2825|M.gilvum_PYR-GCK AAVVGVDDDQARAQPLG----------------RARLRVAGAAGLFVVGF
**:**. * *::*:* ***.**
MMAR_0873|M.marinum_M TTVFVLGTVAVLGMTTTLISNQLVLQRAGGVLTIIMGLVFVGLIPALQRQ
MUL_0626|M.ulcerans_Agy99 TTVFVLGTVAVLGMTTTLISNQLVLQRAGGVLTIIMGLVFVGLIPALQRQ
MAV_4618|M.avium_104 TAVFVLGTVAVLGMTTTLITNQLLLQRAGGVLTIVMGLVFVGLIPALQRQ
Mb0540|M.bovis_AF2122/97 TTVFVLGTVAVLGMTTTLITNQLLLQRVGGVLIVVMGLVFVGFIGALQRQ
Rv0527|M.tuberculosis_H37Rv TTVFVLGTVAVLGMTTTLITNQLLLQRVGGVLIVVMGLVFVGFIGALQRQ
MLBr_02411|M.leprae_Br4923 TTVFVLDTVAVLGMTTVLTTHQVLLQRVGGVLTIVMGLVFVGLLPALQRQ
MSMEG_0972|M.smegmatis_MC2_155 TVVFLLGAVAVLGMTTTLIANQLLLQRIGGVVTIVMGLVFVGFIPALQRQ
TH_2250|M.thermoresistible__bu TVVFLLGTVAVLGMTTTLITNQVLLQRIGGVITIVMGLVFIGFIPALQRQ
MAB_3975c|M.abscessus_ATCC_199 TAVFLLGTVAVMGMTTSLITNQVLLQRIGGVVTIAMGLVFIGLVPALQRD
Mvan_3693|M.vanbaalenii_PYR-1 TAVFLMGTVAILGLTTTLITNGVLLQRIGGVVTIVMGLVFMGFFPALQRD
Mflv_2825|M.gilvum_PYR-GCK TAVFLMGTVAILGLTTTIITNGVLLQRIGGVVTIVMGLVFMGFFPALQRD
*.**::.:**::*:** : :: ::*** ***: : *****:*:. ****:
MMAR_0873|M.marinum_M ARFSPRQVTTVAGAPLLGAVFALGWTPCLGPTLTGVITVASATDGASVAR
MUL_0626|M.ulcerans_Agy99 ARFSPRQVTTVAGAPLLGAVFALGWTPCLGPTLTGVITVASATNGASVAR
MAV_4618|M.avium_104 ARFSPRQLTTVAGAPLLGAVFALGWTPCLGPTLAGVITVASATDGASVAR
Mb0540|M.bovis_AF2122/97 ARFTPRQLTSVAGAPVLGAVFALGWTPCLGPTLTGVITVASATEGASVAR
Rv0527|M.tuberculosis_H37Rv ARFTPRQLTSVAGAPVLGAVFALGWTPCLGPTLTGVITVASATEGASVAR
MLBr_02411|M.leprae_Br4923 VQFSLRQLTTVAGAPVLGTVFALGWTPCLGPTLSGVITVAAATDGVNVTR
MSMEG_0972|M.smegmatis_MC2_155 ARFTPRQLSTVAGAPLLGAVFALGWTPCLGPTLTGVIAVASATDGATVAR
TH_2250|M.thermoresistible__bu ARFAPRQWSTVAGAPLLGAVFALGWTPCLGPTLTGVITVASATDGASVAR
MAB_3975c|M.abscessus_ATCC_199 IRFTPRRVSTVAGAPLLGAVFALGWTPCLGPTLTGVVAVASATEGADVAR
Mvan_3693|M.vanbaalenii_PYR-1 ARFTPTSLSTLWGAPLLGAVFGLGWTPCLGPTLTGVIAVASASEGPNVVR
Mflv_2825|M.gilvum_PYR-GCK ARFTPTSLSTLWGAPLLGAVFGLGWTPCLGPTLTGVIAVASATEGPNVVR
:*: ::: ***:**:**.***********:**::**:*::* *.*
MMAR_0873|M.marinum_M GIVLVIAYCLGLGIPFVLLAFGSAGAVAGLGWLRRHTRAIQIFGGVLLIA
MUL_0626|M.ulcerans_Agy99 GIVLVIAYCLGLGIPFVLLAFGSAGAVAGLGWLRRHTRAIQIFGGVLLIV
MAV_4618|M.avium_104 GIVLVIAYCLGLGIPFVLLAFGSAGAVAGLGWLRRHTRAIQIFGGVLLIT
Mb0540|M.bovis_AF2122/97 GIVLVIAYCLGLGIPFVLLAFGSAWAVAGLGWLRRHTRAIQIFGGALLIA
Rv0527|M.tuberculosis_H37Rv GIVLVIAYCLGLGIPFVLLAFGSAWAVAGLGWLRRHTRAIQIFGGALLIA
MLBr_02411|M.leprae_Br4923 GILLVIAYCMGLGIPFVLLASGSAQAVAGLRWLRQYGRAIQVFGGMLLIA
MSMEG_0972|M.smegmatis_MC2_155 GVVLVLAYCLGLGIPFVLLALGSARAVQGLGWLRRHTRTIQIFGGVLLIL
TH_2250|M.thermoresistible__bu GVVLVIAYCLGLGIPFILLAFGSARAVQGLGWLRRNTRTIQIFGGVLLIL
MAB_3975c|M.abscessus_ATCC_199 GIALVIAYCLGLGMPFLLLAFGSARAVRGMDWLRRHTRMIQVIGGIALLA
Mvan_3693|M.vanbaalenii_PYR-1 GIVLVVAYCVGLGAPFVLFALASGRAMRALGWLRRNARRIQVFGGVLLVA
Mflv_2825|M.gilvum_PYR-GCK GIVLVVAYCVGLGAPFVLFALASGRAMRALGWLRRNARRIQVFGGVLLVA
*: **:***:*** **:*:* .*. *: .: ***: * **::** *:
MMAR_0873|M.marinum_M VGLALVTGVWNDVVSWLRDAFVSDVRLPI
MUL_0626|M.ulcerans_Agy99 VGLALVTGVWNDVVSWLRDAFVSDVRLPI
MAV_4618|M.avium_104 VGALLVTGLWNDFVSWLRDAFVSDVRLPI
Mb0540|M.bovis_AF2122/97 VGAALVTGVWNDVVSWLRDAFVSDVRLPI
Rv0527|M.tuberculosis_H37Rv VGAALVTGVWNDVVSWLRDAFVSDVRLPI
MLBr_02411|M.leprae_Br4923 IGAALIAGVWDDFVSWLRDAVVSDMRVSI
MSMEG_0972|M.smegmatis_MC2_155 VGAALVTGLWNEFVSYVRDAFVSDVRLPI
TH_2250|M.thermoresistible__bu VGAALVTGLWNEFISWVRDAFVSDVRLPI
MAB_3975c|M.abscessus_ATCC_199 VGVALITGVWADFVAWVRDSFVSDARLPI
Mvan_3693|M.vanbaalenii_PYR-1 VGIALVTGLWNEVVSWVRDAFVSNTTLPI
Mflv_2825|M.gilvum_PYR-GCK VGIALLTGLWNEVVSWVRDAFVSNTTLPI
:* *::*:* :.::::**:.**: :.*