For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RRGVRRVRQVLAVAGIAAVVGACASAERGEADRPVVLTTFTVLADIAANVAGEHLVVESITKPGAEIHGY EPTPGDIRKAAQADLILDNGLNLEAWFARFVDGIDVPHVMVSDGIVPLDISADAYAGLPNPHAWMSPLNV QIYADNMATAFSELDPGNAADYAANAAEYRRELQQIHDDLLARLDAVPAAQRALVTCEGAFSYLARDTGL TEKYIWPVNAEQQATPQQIASTIEFVDANDVPAVFCESTVSDAPMQRVVEATGARLGGVLFVDSLSEADG PVPTYLELIRHDAETITAALTGNPKS*
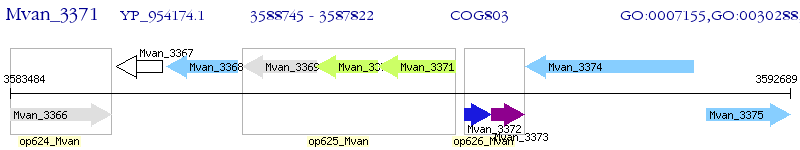
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3371 | - | - | 100% (307) | periplasmic solute binding protein |
| M. vanbaalenii PYR-1 | Mvan_5324 | - | 7e-29 | 28.28% (297) | periplasmic solute binding protein |
| M. vanbaalenii PYR-1 | Mvan_5320 | - | 9e-09 | 20.51% (312) | periplasmic solute binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2085 | - | 2e-14 | 27.90% (233) | hypothetical protein Mb2085 |
| M. gilvum PYR-GCK | Mflv_3168 | - | 1e-137 | 78.10% (306) | periplasmic solute binding protein |
| M. tuberculosis H37Rv | Rv2059 | - | 2e-14 | 27.90% (233) | hypothetical protein Rv2059 |
| M. leprae Br4923 | MLBr_00337 | - | 1e-13 | 24.13% (286) | putative periplasmic solute-binding proteins |
| M. abscessus ATCC 19977 | MAB_0577c | - | 7e-14 | 26.94% (297) | ABC transporter solute-binding protein |
| M. marinum M | MMAR_5071 | - | 1e-14 | 25.42% (299) | hypothetical protein MMAR_5071 |
| M. avium 104 | MAV_0583 | - | 5e-17 | 29.17% (288) | periplasmic solute binding protein family protein |
| M. smegmatis MC2 155 | MSMEG_6050 | - | 2e-30 | 31.53% (295) | solute-binding lipoprotein |
| M. thermoresistible (build 8) | TH_4096 | - | 1e-115 | 71.33% (293) | PUTATIVE periplasmic solute binding protein |
| M. ulcerans Agy99 | MUL_4147 | - | 6e-15 | 26.00% (300) | hypothetical protein MUL_4147 |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00337|M.leprae_Br4923 --------------------------MRWLTTIILCLISSTVAG------
MAV_0583|M.avium_104 -----------MGTALTRDATRVGRTTRWLATVLAVTGWAALTG------
MMAR_5071|M.marinum_M --------------------MLRVRVAHGFSAILLSVLGATGVA------
MUL_4147|M.ulcerans_Agy99 ---------------------MRVRVAHGFSAILLSVLGATGVA------
Mb2085|M.bovis_AF2122/97 MATPVILVTGHEGTAAVTADLLGLLTDHGTATLRSVAPGSVRRADPRPRC
Rv2059|M.tuberculosis_H37Rv MATPVILVTGHEGTAAVTADLLGLLTDHGTATLRSVAPGSVRRADPRPRC
MAB_0577c|M.abscessus_ATCC_199 ----------------------MPARVAGLAVAVATAGTLALSG------
Mvan_3371|M.vanbaalenii_PYR-1 ------------------MRRGVRRVR----QVLAVAGIAAVVG------
Mflv_3168|M.gilvum_PYR-GCK ----------------MGARHASRRARGRPATWVAIACCASMVA------
TH_4096|M.thermoresistible__bu --------------------MEIRRCLG---TVLAVWALVFGLL------
MSMEG_6050|M.smegmatis_MC2_155 ---------------------MTRRCIAACLAILLLAACGAPDG------
MLBr_00337|M.leprae_Br4923 -------------GCGSLSSGHLR---------------PTAVVAST---
MAV_0583|M.avium_104 -------------CTG---PAHPH---------------ATAVVAST---
MMAR_5071|M.marinum_M -------------ACGTDHDAHPG---------------AAAVVAST---
MUL_4147|M.ulcerans_Agy99 -------------ACGTDHDAHPG---------------AAAVVAST---
Mb2085|M.bovis_AF2122/97 HRREQRRRHRASMKSAIHPDHHPRRLPRCPVLRRDQVVLEMIVITMVGRP
Rv2059|M.tuberculosis_H37Rv HRREQRRRHRASMKSAIHPDHHPRRLPRCPVLRRDQVVLEMIVITMVGRP
MAB_0577c|M.abscessus_ATCC_199 --------------CGQSHERSG----------------ALTVVAST---
Mvan_3371|M.vanbaalenii_PYR-1 -------------ACASAERGEADR--------------PVVLTTFT---
Mflv_3168|M.gilvum_PYR-GCK -------------CSSPIDTGDAAR--------------PTVLTTFT---
TH_4096|M.thermoresistible__bu -------------GCGPHRGAGDDT--------------PMVLTTFT---
MSMEG_6050|M.smegmatis_MC2_155 ---------------SDGRR--------------------QIVVTTT---
. : : .
MLBr_00337|M.leprae_Br4923 --------DVWGSVARIIAGRHVAVTSIVTGAHTDPHTYRVNPAETAAIT
MAV_0583|M.avium_104 --------DVWGSVARAVAGGHVAVASILSGSDQDPHSYEASPSDAAAIA
MMAR_5071|M.marinum_M --------DVWGSVARAVAGGHLAVKSILTGAASDPHSYQASPVDAAMIA
MUL_4147|M.ulcerans_Agy99 --------DVWGSVARAVAGGHLAVKSILTGAASDPHSYQASPVDAAMIA
Mb2085|M.bovis_AF2122/97 SGPGERKWDVWGSVARAVTGGHVPVKSILTGAHADPHSYQASPADAAAIV
Rv2059|M.tuberculosis_H37Rv SGPGERKWDVWGSVARAVTGGHVPVKSILTGAHADPHSYQASPADAAAIV
MAB_0577c|M.abscessus_ATCC_199 --------DAWASVAQAVTGDHAKVSALVSGADTDPHSFEVTPAAAAQVQ
Mvan_3371|M.vanbaalenii_PYR-1 ---------VLADIAANVAGEHLVVESITKPG-AEIHGYEPTPGDIRKAA
Mflv_3168|M.gilvum_PYR-GCK ---------VLADIAANVAGDHLTVESITKPG-AEIHGYEPTPGDIRKAA
TH_4096|M.thermoresistible__bu ---------VLADIARNVAGEHLQVESITKVG-AEIHGYEPTPGDIRKAA
MSMEG_6050|M.smegmatis_MC2_155 ---------ILGDVVSEVVGDTADVRVMMKPN-ADPHSYGLSAADAAEMA
..:. :.* * : . : * : ..
MLBr_00337|M.leprae_Br4923 DAALVVYNGGGYDPWVDKVLAGR-----PDIKSVDAYSLLASRGATTEGP
MAV_0583|M.avium_104 DAGLVVFNGGGYDGWVDDVLAHH-----PGVARVDAYALLPDAGR----P
MMAR_5071|M.marinum_M DSSLLIYNGGGYDPWVDNVLANH-----PAINAIDAYSLLGTDAAR---P
MUL_4147|M.ulcerans_Agy99 DSSLLIYNGGGYDPWVDNVLANH-----PAINAIDAYPLLGTDAAR---P
Mb2085|M.bovis_AF2122/97 DAELVIYNGGGYDPWVDQVLAGH-----PGVQAVDAYSLLGAVGDDD--A
Rv2059|M.tuberculosis_H37Rv DAELVIYNGGGYDPWVDQVLAGH-----PGVQAVDAYSLLGAVGDDD--A
MAB_0577c|M.abscessus_ATCC_199 DATLVVYNGDGYDPFIDKLLKD-------GEPRVNAYQLLPADSAD----
Mvan_3371|M.vanbaalenii_PYR-1 QADLILDNGLNLEAWFARFVDGIDV---PHVMVSDGIVPLDISADAYAGL
Mflv_3168|M.gilvum_PYR-GCK RADLILDNGLNLEAWFARFVDGIDV---PHVVVSDGVAPLEIAEDAYTGM
TH_4096|M.thermoresistible__bu RADLILDNGLNLEAWFEQFISEVNV---ERVRVSDGIEPIGIAG---TDQ
MSMEG_6050|M.smegmatis_MC2_155 RADLVIYNGLGLEESMQRHVDAAAENGVRTLPVGDHVDPLQFSADEHG-A
: *:: ** . : . : : :
MLBr_00337|M.leprae_Br4923 ADEHVFYDLNIAKSVASLIADQLVTIDPDNAADYQANATEFCRSADAIAI
MAV_0583|M.avium_104 RNEHVFYHLGVAKAVAAAVADRLAAIDPGNAADYRRNAAAFGRDADAIAG
MMAR_5071|M.marinum_M PDEHVFYNLHVAKSVAATIAERLAIIDPAHAAEYRTNAVEFGRNADAIVD
MUL_4147|M.ulcerans_Agy99 PDEHVFYNLHVAKSVAATIAERLAIIDPAHAAEYRTNAVEFGRNADAIGD
Mb2085|M.bovis_AF2122/97 PNEHVFYDPNVAKAVAATIADRLADLDPSNSGNYRANAAEFSRGADAIAI
Rv2059|M.tuberculosis_H37Rv PNEHVFYDPNVAKAVAATIADRLADLDPSNSGNYRANAAEFSRGADAIAI
MAB_0577c|M.abscessus_ATCC_199 KNEHVFYSLRTAQQVANNVADDLAKADPGNADSYHANAKTFGQQLDSIAS
Mvan_3371|M.vanbaalenii_PYR-1 PNPHAWMSPLNVQIYADNMATAFSELDPGNAADYAANAAEYRRELQQIHD
Mflv_3168|M.gilvum_PYR-GCK ANPHAWMSPLNVQIYADNMADAFADLDPDHAQAYRDNAAAYRTQLQEVHD
TH_4096|M.thermoresistible__bu PNPHAWMSPLNVITYVDNMADAFSALDPANAEHYRANAEAYQQELRAVHE
MSMEG_6050|M.smegmatis_MC2_155 ADPHFWTDPQRMILAVDAITGRVAELPGIDRAAVAANAARYRDELNNLSA
: * : . :: . . ** : :
MLBr_00337|M.leprae_Br4923 SEHAIASDYPAA--GVIVTEPVVHYLLQASGLVNRTPPAFTATHENENDP
MAV_0583|M.avium_104 IEHTIAAAHPGG--SVVATEPVAFYLLEASGLVNRTPPALEAAVENETDP
MMAR_5071|M.marinum_M SERAIGAEHPAV--GVIATEPVVHYLLAASGLVNRTPAAFAQANENDSDP
MUL_4147|M.ulcerans_Agy99 SERAIGAEHPAV--GVIATEPVVHYLLAASGLVNRTPAAFAQANENDSDP
Mb2085|M.bovis_AF2122/97 SEHAIATTYPDA--AVIATEPVVHYLLAAAGLKNRTPATFIAANENGNDP
Rv2059|M.tuberculosis_H37Rv SEHAIATTYPDA--AVIATEPVVHYLLAAAGLKNRTPATFIAANENGNDP
MAB_0577c|M.abscessus_ATCC_199 LQEDVATKYRGK--AILATEPVASHLVSRCGLVDRTPHAFAEAAEQGQDP
Mvan_3371|M.vanbaalenii_PYR-1 DLLARLDAVPAAQRALVTCEGAFSYLARDTGLTEK----YIWPVNAEQQA
Mflv_3168|M.gilvum_PYR-GCK ELVERLDALPPTQRALVTCEGAFSYLARDAGLTEK----YIWAVNAEQQA
TH_4096|M.thermoresistible__bu DLVNRLSGLPPGQRALVSCEGAFSYLARDAGLTEK----YIWPVNAEQQA
MSMEG_6050|M.smegmatis_MC2_155 HMTDRFAAIPAQRRKLVTNHHVLGYLADRFGFTVIG--AVIPSGTTLASP
:: . . :* *: . ..
MLBr_00337|M.leprae_Br4923 SAADMAAALNLINHRQVSALLVNPQKSNAATNGLQAAARRSGVPVTEVTE
MAV_0583|M.avium_104 APADLARALDLLDRRQVSALVVNPQTSASAVNGLREAARRAGVPVVEVRE
MMAR_5071|M.marinum_M APADLASVLTLIDRREVAALLVNPQTATATTAGLQAAAHRAGVPVAEVSE
MUL_4147|M.ulcerans_Agy99 APADLASVLTLIDRREVAALLVNPQTATATTAGLQAAAHRAGVPVAEVSE
Mb2085|M.bovis_AF2122/97 TPADMAAVLDMIAGREVAALLVNPQTPTAATDELQVAARRAGVPITELTE
Rv2059|M.tuberculosis_H37Rv TPADMAAVLDMIAGREVAALLVNPQTPTAATDELQVAARRAGVPITELTE
MAB_0577c|M.abscessus_ATCC_199 SPADVASALDLISTKQVAALLFNPQTAGSVTNRIKQAAESHGVPVVTVTE
Mvan_3371|M.vanbaalenii_PYR-1 TPQQIASTIEFVDANDVPAVFCESTVSDAPMQRVVEATGARLGGVLFVDS
Mflv_3168|M.gilvum_PYR-GCK TPQQIASTIEFVDANDVPAVFCESTVSDAPMQRVVEATGTRFGGVLFVDS
TH_4096|M.thermoresistible__bu TPRQIASVIEFVNANAVPAVFCESTVSDAPMQRVVEATGAVFGGVLYVDS
MSMEG_6050|M.smegmatis_MC2_155 SASDLESLAGAIRDAGVKAIFVDSSQPDRLARVLAEQSGVRVNVVSLYSE
:. :: : * *:. :. . : : : .
MLBr_00337|M.leprae_Br4923 MLPNDTDY------------------------LTWQRNTIDQLL-TALQS
MAV_0583|M.avium_104 TLPDGADY------------------------LSWQRNTVGQLQ-TALQP
MMAR_5071|M.marinum_M TMPIDTDY------------------------LTWQRNTVDQLA-AALRS
MUL_4147|M.ulcerans_Agy99 TMPIDTDY------------------------LTWQRNTVDQLA-AALRS
Mb2085|M.bovis_AF2122/97 TLPSGTDRDQFCAADRPDRRGRSLRADHADRGLSARGHRVGDLLPTALVC
Rv2059|M.tuberculosis_H37Rv TLPSGTDRDQFCAADRPDRRGRSLRADHADRGLSARGHRVGDLLPTALVC
MAB_0577c|M.abscessus_ATCC_199 TLPEGTDY------------------------VTWQRRTTEQLA-AALG-
Mvan_3371|M.vanbaalenii_PYR-1 LSEADGPVP---------------------TYLELIRHDAETIT-AALTG
Mflv_3168|M.gilvum_PYR-GCK LSEADGPVP---------------------TYLDLIRHDATTIA-AALTA
TH_4096|M.thermoresistible__bu LSEPDGPVP---------------------TYLDLIRHDADVIA-AGLAG
MSMEG_6050|M.smegmatis_MC2_155 SLSAPGTEADS--------------------YLGMMRTNTETIANNLI--
: : :
MLBr_00337|M.leprae_Br4923 NRSPR---------------------------------------------
MAV_0583|M.avium_104 VRSLQP--------------------------------------------
MMAR_5071|M.marinum_M GR------------------------------------------------
MUL_4147|M.ulcerans_Agy99 GR------------------------------------------------
Mb2085|M.bovis_AF2122/97 HRRSGGRGRPRRASARPGNCVRRTDGRGSRPGCPDRRGTPRDVFADHPRR
Rv2059|M.tuberculosis_H37Rv HRRSGGRGRPRRASARPGNCVRRTDGRGSRPGCPDRRGTPRDVFADHPRR
MAB_0577c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_3371|M.vanbaalenii_PYR-1 NPKS----------------------------------------------
Mflv_3168|M.gilvum_PYR-GCK DRAS----------------------------------------------
TH_4096|M.thermoresistible__bu RRS-----------------------------------------------
MSMEG_6050|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00337|M.leprae_Br4923 --------------------------------------------------
MAV_0583|M.avium_104 --------------------------------------------------
MMAR_5071|M.marinum_M --------------------------------------------------
MUL_4147|M.ulcerans_Agy99 --------------------------------------------------
Mb2085|M.bovis_AF2122/97 GGRPGRGCPGRRDRDLGGLRRGFRRRRHPAVAGAWSPGVGVRGHHLVCDL
Rv2059|M.tuberculosis_H37Rv GGRPGRGCPGRRDRDLGGLRRGFRRRRHPAVAGAWSPGVGVRGHHLVCDL
MAB_0577c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_3371|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3168|M.gilvum_PYR-GCK --------------------------------------------------
TH_4096|M.thermoresistible__bu --------------------------------------------------
MSMEG_6050|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00337|M.leprae_Br4923 --------------------
MAV_0583|M.avium_104 --------------------
MMAR_5071|M.marinum_M --------------------
MUL_4147|M.ulcerans_Agy99 --------------------
Mb2085|M.bovis_AF2122/97 PDLLVAPAAPLTSRSRFRPL
Rv2059|M.tuberculosis_H37Rv PDLLVAPAAPLTSRSRFRPL
MAB_0577c|M.abscessus_ATCC_199 --------------------
Mvan_3371|M.vanbaalenii_PYR-1 --------------------
Mflv_3168|M.gilvum_PYR-GCK --------------------
TH_4096|M.thermoresistible__bu --------------------
MSMEG_6050|M.smegmatis_MC2_155 --------------------