For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAILGYARVSTLGQDLDAQLAALAAQGVESGRIFADKLSGAVNTVRPGLAALLHYAREGDIVVVTAIDRL GRSVAEVTRTIADLGERRILLRALREGVDTGTPTGRAVATIMATLAELELELGRERRAASRDSRRARQLP ATKPAKLTPERQEQLRRLAGTGEPVHELAQAFGVSRATAYRYLAKLSFQSGVA*
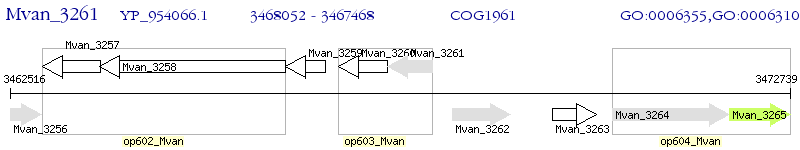
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3261 | - | - | 100% (194) | resolvase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_4555 | - | 8e-32 | 58.33% (120) | resolvase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1448 | - | 5e-07 | 34.04% (94) | recombinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1612c | - | 2e-06 | 32.35% (102) | phiRv1 integrase |
| M. gilvum PYR-GCK | Mflv_2148 | - | 1e-29 | 54.17% (120) | resolvase domain-containing protein |
| M. tuberculosis H37Rv | Rv1586c | - | 2e-06 | 32.35% (102) | phiRv1 integrase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1820c | - | 1e-48 | 57.07% (184) | putative resolvase |
| M. marinum M | MMAR_0589 | pinR | 8e-51 | 55.50% (191) | site-specific recombinase PinR |
| M. avium 104 | MAV_1493 | - | 8e-09 | 31.61% (155) | Y4cG protein |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3340 | - | 1e-22 | 35.52% (183) | PUTATIVE putative Resolvase, N-terminal domain |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3261|M.vanbaalenii_PYR-1 ----------------MTAILGYARVSTLGQD--LDAQLAALAAQGVESG
Mflv_2148|M.gilvum_PYR-GCK ----------------MAVTLGYATAVPGCAD--LDDQLAALTAAGVDPR
MAB_1820c|M.abscessus_ATCC_199 --------MDTSTTADTGVLLGYARVSTAHQS--LDQQLDALTEAGVGPD
MMAR_0589|M.marinum_M ----------MTAPATSGTLLGYARVSTGHQS--LDQQLDALSAEGVATE
TH_3340|M.thermoresistible__bu ---------------MTGQLVGYIRVSTLDQN--TERQLD-----GIDLD
Mb1612c|M.bovis_AF2122/97 --------MRYTTPVRAAVYLRISEDRSGEQLGVARQREDCLKLCGQRKW
Rv1586c|M.tuberculosis_H37Rv --------MRYTTPVRAAVYLRISEDRSGEQLGVARQREDCLKLCGQRKW
MAV_1493|M.avium_104 MGMTAPRTRRRTQTAPAGTVVAYVRVSTEEQAASGAGLDAQRAAIAAECD
: . .
Mvan_3261|M.vanbaalenii_PYR-1 R-------IFADK-LSG-AVNTVRPGLAALLHYAREG--DIVVVTAIDRL
Mflv_2148|M.gilvum_PYR-GCK R-------VFTDR-TPG-SADKMRAGMLALLSYARAG--DVVVVAALERL
MAB_1820c|M.abscessus_ATCC_199 R-------IYKDK-LSGTSTREQRPGLAALLDYARPG--DGIVVVGIDRL
MMAR_0589|M.marinum_M R-------IYQDK-LSGMSTRDQRPGLAALLDYARPG--DTITVVGIDRL
TH_3340|M.thermoresistible__bu K-------VFTDK-ASG--KDTNRPALDECLRYVREG--DTLIVHSMDRL
Mb1612c|M.bovis_AF2122/97 VP-----VEYLDN-DVSASTGKRRPAYEQMLADITAGKIAAVVAWDLDRL
Rv1586c|M.tuberculosis_H37Rv VP-----VEYLDN-DVSASTGKRRPAYEQMLADITAGKIAAVVAWDLDRL
MAV_1493|M.avium_104 RRGWTVVGWYADEGVSGGKALDARPALTAALNTVETAQAAVLMVAKSDRL
: *. . *.. * . : . :**
Mvan_3261|M.vanbaalenii_PYR-1 GRSVAEVTRTIADLGERRILLRALREGV----DTGTPTGRAVATIMATLA
Mflv_2148|M.gilvum_PYR-GCK GRSAAEVTRTVADLTTRGITLRCLADGL----DTATSTGRVVAKVLTDLA
MAB_1820c|M.abscessus_ATCC_199 GRNAAEVMATIRDLAEREIVLHSLREGI----DSSNATGRMIAGVLASLA
MMAR_0589|M.marinum_M GRNAAEVMTTIRDLSQRGIVLRSLREGI----DTSNATGRMVAGVLASLA
TH_3340|M.thermoresistible__bu ARSLPDLRRLVDELTARGVAVRFVKEGLTFTRDESDPCSVLMLSVMGAVA
Mb1612c|M.bovis_AF2122/97 HRRPIELEAFMSLADEKRLALATVAGDV----DLATPQGRLVARLKGSVA
Rv1586c|M.tuberculosis_H37Rv HRRPIELEAFMSLADEKRLALATVAGDV----DLATPQGRLVARLKGSVA
MAV_1493|M.avium_104 ARSLPTLLHVIDRAERAGGVVVAVDGTV----DTSTAAGRFTTQIMGSVA
* : : : : : * . . . : :*
Mvan_3261|M.vanbaalenii_PYR-1 ELELELGRERRAASRDSRRARQLP-------------ATKPAKLTPERQE
Mflv_2148|M.gilvum_PYR-GCK SLD-----------------------------------------APDSAE
MAB_1820c|M.abscessus_ATCC_199 ELELELGRERRAAARESRRARGQH-------------IGRPKALDNAKAA
MMAR_0589|M.marinum_M ELELELGRERRSAAREARRSRGQS-------------IGRPKALDESKAA
TH_3340|M.thermoresistible__bu EFERSLMLQRQREGIAIAKAKGVY-------------KGRKPALSDAQAA
Mb1612c|M.bovis_AF2122/97 AHETEHKKARQRRAARQKAERGHPNWSKAFGYLPGPNGPEPDPRTAPLVK
Rv1586c|M.tuberculosis_H37Rv AHETEHKKARQRRAARQKAERGHPNWSKAFGYLPGPNGPEPDPRTAPLVK
MAV_1493|M.avium_104 ELERGLISDRTKAALAVRKAQGVR-------------LGRPTTVPSSVVD
:
Mvan_3261|M.vanbaalenii_PYR-1 QLRRLAGTGEPVHELAQAFGVS------------RATAYRYLAKLSFQSG
Mflv_2148|M.gilvum_PYR-GCK SAHRA---------------------------------------------
MAB_1820c|M.abscessus_ATCC_199 LAQRMHLAGEPVLTIANTLGVS------------RATVYRVLAEDLHAGK
MMAR_0589|M.marinum_M LARRMHAAGEPVSTIGSALGVS------------RATVYRVLAETEGEAA
TH_3340|M.thermoresistible__bu EVAQRLSAGESATELAREYGVS------------RATVYNVRNRARGAA-
Mb1612c|M.bovis_AF2122/97 QAYADILAGASLGDVCRQWNDAGAFT-ITGRPWTTTTLSKFLRKPRNAGL
Rv1586c|M.tuberculosis_H37Rv QAYADILAGASLGDVCRQWNDAGAFT-ITGRPWTTTTLSKFLRKPRNAGL
MAV_1493|M.avium_104 RIAAERAAGATWAAIADGLNADGTPTGQGGSQWFPNTVARIAKRAVAVPA
Mvan_3261|M.vanbaalenii_PYR-1 VA------------------------------------------------
Mflv_2148|M.gilvum_PYR-GCK --------------------------------------------------
MAB_1820c|M.abscessus_ATCC_199 EDQ-----------------------------------------------
MMAR_0589|M.marinum_M V-------------------------------------------------
TH_3340|M.thermoresistible__bu --------------------------------------------------
Mb1612c|M.bovis_AF2122/97 RAYKGARYGPVDRDAIVGKAQWSPLVDEATFWAAQAVLDAPGRAPGRKSV
Rv1586c|M.tuberculosis_H37Rv RAYKGARYGPVDRDAIVGKAQWSPLVDEATFWAAQAVLDAPGRAPGRKSV
MAV_1493|M.avium_104 T-------------------------------------------------
Mvan_3261|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2148|M.gilvum_PYR-GCK --------------------------------------------------
MAB_1820c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0589|M.marinum_M --------------------------------------------------
TH_3340|M.thermoresistible__bu --------------------------------------------------
Mb1612c|M.bovis_AF2122/97 RRHLLTGLAGCGKCGNHLAGSYRTDGQVVYVCKACHGVAILADNIEPILY
Rv1586c|M.tuberculosis_H37Rv RRHLLTGLAGCGKCGNHLAGSYRTDGQVVYVCKACHGVAILADNIEPILY
MAV_1493|M.avium_104 --------------------------------------------------
Mvan_3261|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2148|M.gilvum_PYR-GCK --------------------------------------------------
MAB_1820c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0589|M.marinum_M --------------------------------------------------
TH_3340|M.thermoresistible__bu --------------------------------------------------
Mb1612c|M.bovis_AF2122/97 HIVAERLAMPDAVDLLRREIHDAAEAETIRLELETLYGELDRLAVERAEG
Rv1586c|M.tuberculosis_H37Rv HIVAERLAMPDAVDLLRREIHDAAEAETIRLELETLYGELDRLAVERAEG
MAV_1493|M.avium_104 --------------------------------------------------
Mvan_3261|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2148|M.gilvum_PYR-GCK --------------------------------------------------
MAB_1820c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0589|M.marinum_M --------------------------------------------------
TH_3340|M.thermoresistible__bu --------------------------------------------------
Mb1612c|M.bovis_AF2122/97 LLTARQVKISTDIVNAKITKLQARQQDQERLRVFDGIPLGTPQVAGMIAE
Rv1586c|M.tuberculosis_H37Rv LLTARQVKISTDIVNAKITKLQARQQDQERLRVFDGIPLGTPQVAGMIAE
MAV_1493|M.avium_104 --------------------------------------------------
Mvan_3261|M.vanbaalenii_PYR-1 --------------------------------------
Mflv_2148|M.gilvum_PYR-GCK --------------------------------------
MAB_1820c|M.abscessus_ATCC_199 --------------------------------------
MMAR_0589|M.marinum_M --------------------------------------
TH_3340|M.thermoresistible__bu --------------------------------------
Mb1612c|M.bovis_AF2122/97 LSPDRFRAVLDVLAEVVVQPVGKSGRIFNPERVQVNWR
Rv1586c|M.tuberculosis_H37Rv LSPDRFRAVLDVLAEVVVQPVGKSGRIFNPERVQVNWR
MAV_1493|M.avium_104 --------------------------------------