For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTDVTNPPADPIEPPAAKPPVLAWWTRGDTNAFFGLGFNILVNVLTLTGLMIGVVAVPADDVLGTVLPA LGVALILGNLYYTFLARRLAKRENRSDVTALPYGPSVPHMFIVVFVVMLPVYLATDDPTQAWQAGLAWAF LIGVIVMIGAFVGPYIRKLTPRAAMLGTLAGISITFISMRPAAQMWEVAWIGLPVLAIILIGFFTDVKLP GGVPIGLVALLVGTAIGWAGGYMSAPDVGQAFSDIAIGIPDLRIDMLFNGLADLAPLLGTAIPLGVYNFT EAMSNVESAAAAGDNYNLRSVLLADGAGAVIGSAFGSPFPPAVYIGHPGWKDAGGRAGYSLASGAVIGIF CFVGLFGVLAALLPVPAIVPILLYIGLLIGAQAFQAVPRLHAVAVVAALLPNLAEWGRGMIDNALSAAGT SAAEVTADALNGAGVVYEGLKTLGEGAVLVGLILGTMVTYILDKKFLHAAAASAVGAALSFIGLIHAPEV AWAASPQVALGYLLFGGVCVAFWFLPGAKEPVVVDEADIVAGH
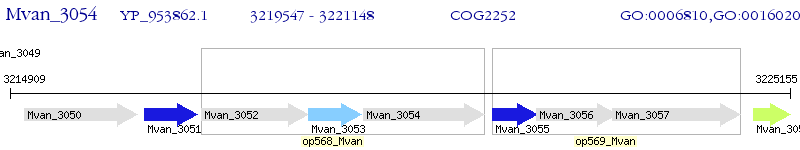
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3054 | - | - | 100% (533) | xanthine/uracil/vitamin C permease |
| M. vanbaalenii PYR-1 | Mvan_1159 | - | 1e-08 | 23.94% (284) | sulphate transporter |
| M. vanbaalenii PYR-1 | Mvan_1430 | - | 7e-07 | 21.67% (443) | sulfate transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4975 | - | 5e-10 | 23.44% (418) | sulfate transporter |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3353c | - | 7e-08 | 25.43% (405) | putative xanthine/uracil/vitamin C permease family protein |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1448 | - | 7e-08 | 22.22% (279) | integral membrane transporter |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4975|M.gilvum_PYR-GCK -----------MTSNFGELRSYRRSALRDDTQAGLSVAAYLVPQALAYAT
MSMEG_1448|M.smegmatis_MC2_155 -MTSATITPREEQSVWAALRSPRR--LRTEVLAGLVVALALIPEAISFSI
Mvan_3054|M.vanbaalenii_PYR-1 MSTDVTNPPADPIEPPAAKPPVLAWWTRGDTNAFFGLGFNILVNVLTLTG
MAB_3353c|M.abscessus_ATCC_199 -----MTALLDRFFRITERQSTLGREIRGGVVTFFAMAYIVVLNPLIIGG
. * . : : :. :: : :
Mflv_4975|M.gilvum_PYR-GCK LAG---LSPAAGLWAALPPLLVYAILGS-------------------SRQ
MSMEG_1448|M.smegmatis_MC2_155 IAG---VDPRVGLFASFTMAVTIAIVGG-------------------RPA
Mvan_3054|M.vanbaalenii_PYR-1 LMIGVVAVPADDVLGTVLPALGVALILGNLYYTFLARRLAKRENRSDVTA
MAB_3353c|M.abscessus_ATCC_199 EPG---RAHNVDVLGHVLPVGQVAAVTALAAG---------------VMS
.: . . * : .
Mflv_4975|M.gilvum_PYR-GCK LSVGPESTTALMTAAVLAP--VVGGDDPVRYAASAAVLAILVGLVCLGAG
MSMEG_1448|M.smegmatis_MC2_155 MISAATGAVALVVAPLVR-------SHGLDYLIATVILAGLLQLVLGGLG
Mvan_3054|M.vanbaalenii_PYR-1 LPYGPSVPHMFIVVFVVMLPVYLATDDPTQAWQAGLAWAFLIGVIVMIGA
MAB_3353c|M.abscessus_ATCC_199 ILFGFVANYPFALAAGLGIN-----SLLAVSFAPQMTWPEAMGLVIVDG-
: . : . : . . . : ::
Mflv_4975|M.gilvum_PYR-GCK MLRLGYLANMLSKPVLVGYMLGVALAMVAGQLGRIAGVEVTGDTVPAQLA
MSMEG_1448|M.smegmatis_MC2_155 VARL---MRFVPRSVMVGFVNALAILIFLAQIPHLL--------------
Mvan_3054|M.vanbaalenii_PYR-1 FVGP-YIRKLTPRAAMLGTLAGISITFISMRPAAQMW-------------
MAB_3353c|M.abscessus_ATCC_199 ------------LIIVALGISGFRTAVFHAIPDELK--------------
: : .. ..
Mflv_4975|M.gilvum_PYR-GCK AFAAAVDGVHWPTAGLALGVLVLVLTMERVAPTLPAPLIGVVVATVVTWA
MSMEG_1448|M.smegmatis_MC2_155 -------GVPWLVYPMVAVALVIIVALPKLTAAVPGPLVAIVLLTAATIG
Mvan_3054|M.vanbaalenii_PYR-1 -------EVAWIGLPVLAIILIGFFTDVKLPGGVPIGLVALLVGTAIGWA
MAB_3353c|M.abscessus_ATCC_199 ---------AAIAAGIGCFIAFIGFVDAGFVRRVPD-AANTTVPVGLGID
: . .. . :* : .
Mflv_4975|M.gilvum_PYR-GCK GELT-RFGVAVV-GDVPVGLPAPALP--WLTVDQIQALIVPAIGISVVAF
MSMEG_1448|M.smegmatis_MC2_155 FGWS-VPDVADE-GRLPSSLPSLLIPNVPLNLDTLKIIAPYALTMAVVGL
Mvan_3054|M.vanbaalenii_PYR-1 GGYMSAPDVGQAFSDIAIGIPDLRIDMLFNGLADLAPLLGTAIPLGVYNF
MAB_3353c|M.abscessus_ATCC_199 NSVATIPTLIFAAGVLLMGILVVRKVRGGLLIGIVAMTVVSMIAQALCGR
: . : .: : : : .:
Mflv_4975|M.gilvum_PYR-GCK SDNILTARAFASRAGQNIDPNTELRALGVSNLAVGFLRGFPVSSSASRTA
MSMEG_1448|M.smegmatis_MC2_155 LESLMTAKLVDDITDTHSNKSREALGQGAANIVTGFFGGMGGCAMIGQTM
Mvan_3054|M.vanbaalenii_PYR-1 TEAMSNVESAAAAGDNYNLRSVLLADGAGAVIGSAFGSPFPPAVYIGHPG
MAB_3353c|M.abscessus_ATCC_199 GAQPTDAKGWNLTVPDWPSSAGGMPDLSLLGQVDLFG-AFTRVGVLSATV
.. . * : . .
Mflv_4975|M.gilvum_PYR-GCK L-AAAAGARTQMYSLVVLAVVAVVVVFGGGLVSHVPAAALGGLIVYAALK
MSMEG_1448|M.smegmatis_MC2_155 INVKACGARTRISTFSAGAFLLGLVVGLGDIVGRIPMAALVAVMVMVSVG
Mvan_3054|M.vanbaalenii_PYR-1 WKDAGGRAGYSLASGAVIGIFCFVGLFG-VLAALLPVPAIVPILLYIGL-
MAB_3353c|M.abscessus_ATCC_199 LVFALVLSNFFDAMGTMTGLGKEAGLAD--EHGTLPGIGRALTVEGVGAV
: .. : . :* . : .
Mflv_4975|M.gilvum_PYR-GCK LVDVRSFR--ALARFRRSE-VVLAALTAIAVTVFGLLYGVVIAVALSVLD
MSMEG_1448|M.smegmatis_MC2_155 TFDWHSINPKTLRRMPKSETTVMLATVAVTVATGNLAYGVAVGTLTAMVL
Mvan_3054|M.vanbaalenii_PYR-1 LIGAQAFQ--AVPRLHAVAVVAALLPNLAEWGRGMIDNALSAAGTSAAEV
MAB_3353c|M.abscessus_ATCC_199 IGGVSSSSSNTVYVESASGIAEGARTGLANVVTGLLFLAAMFFTPLYSVV
. : :: . : .
Mflv_4975|M.gilvum_PYR-GCK LLRRLSHAHDSVQGLVPGLAGMHDVDDYPDARIVPGLVVYRYDAPLCFAN
MSMEG_1448|M.smegmatis_MC2_155 FARRVAHLTEVTTVGQP----------------DPDTRVYAVRGELFFAS
Mvan_3054|M.vanbaalenii_PYR-1 TADALNGAGVVYEGLKT---------------LGEGAVLVGLILGTMVTY
MAB_3353c|M.abscessus_ATCC_199 PLEAAAPALVVVGAMMIG------------QLRSVDLTRFDYALPCFLTV
. .:
Mflv_4975|M.gilvum_PYR-GCK AEDFRRRALEAIDQMDQPTRWFVLNAEANVEVDMTAIEALDQVRQDCLRR
MSMEG_1448|M.smegmatis_MC2_155 SNDL----VYQFDYVGDPD-NIVIDMSQAHIWDASTVATLDAITTKYAAK
Mvan_3054|M.vanbaalenii_PYR-1 ILDKKFLHAAAASAVGAALSFIGLIHAPEVAWAASPQVALGYLLFG----
MAB_3353c|M.abscessus_ATCC_199 VTMPFTYSIANGIGVGFISWVVLALARGRFRSVHPLLYVVAMLFVIYFAK
:. . . .: :
Mflv_4975|M.gilvum_PYR-GCK GIVFAMARVKQDLREALAAGGVLDEIGQDRLYMTLPTAVAAYQAWVASGN
MSMEG_1448|M.smegmatis_MC2_155 GKTVRIIGMNDNS-----------AQRHARLSGQLTGAH-----------
Mvan_3054|M.vanbaalenii_PYR-1 GVCVAFWFLPG---------------AKEPVVVDEADIVAGH--------
MAB_3353c|M.abscessus_ATCC_199 GPIEDLLAHGA---------------------------------------
* :
Mflv_4975|M.gilvum_PYR-GCK KG
MSMEG_1448|M.smegmatis_MC2_155 --
Mvan_3054|M.vanbaalenii_PYR-1 --
MAB_3353c|M.abscessus_ATCC_199 --