For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGRRLRVVIIGGGFGGLFCARRLARADVDVTVLDRTAGHLFQPLLYQCATGTLSIGHISRPLREEFAGHP NITTLLGEAVALDPERRTVTALRPDETTFTIDYDVLVVAAGMQQSYFGKPHFAQWAPGMKTLDDALSIRQ RIFTAFEIAETLPPGAERDAWLTFAVTGGGPTGVELAGQIREMATRTLTNEFHSIEPEEARVLLFDGGDR VLKSFAPELSGRAAGTLRSLGVDVRLGVHVTDVQREGVTVAPKSGGSEEYYAVRTVLWTAGVEAVPFAGH LAEVLDVQADRAGRLAVQPDLSVPGHPEIFVIGDLVGRDDLPGVAENAMQGGLHVAACIRRDLAGRPRRN YRYRDLGSAAYISRGNALLQVGPVKVAGFLGWIAWGLLHIAFLTGVRNRVSTVVTWLATIARASRYHRAF MLGSVSAPEARYTWSSRDQPTPPQALCEDPRTGHTSHMTISAQDVRRLLDAEHDSTLVLIEGRTEVITGQ QKSSDQYRGAFEVISRDELLSRTGGAAHLTDDQISAQAAALDTAVNELGG*
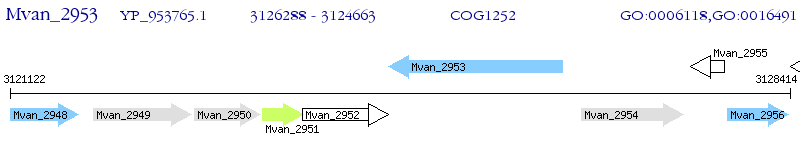
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2953 | - | - | 100% (541) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_3078 | - | 2e-97 | 45.30% (415) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5760 | - | 7e-18 | 29.94% (354) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1885c | ndh | 9e-99 | 45.91% (416) | NADH dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3459 | - | 0.0 | 73.44% (433) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. tuberculosis H37Rv | Rv1854c | ndh | 6e-99 | 45.91% (416) | NADH dehydrogenase |
| M. leprae Br4923 | MLBr_02061 | - | 1e-92 | 43.99% (416) | putative dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2429c | - | 1e-101 | 46.51% (415) | NADH dehydrogenase (NDH) |
| M. marinum M | MMAR_2728 | ndh | 1e-102 | 47.23% (415) | NADH dehydrogenase Ndh |
| M. avium 104 | MAV_2867 | - | 1e-99 | 46.39% (416) | DoxD family protein/pyridine nucleotide-disulfide |
| M. smegmatis MC2 155 | MSMEG_3457 | - | 0.0 | 77.95% (440) | DoxD family protein/pyridine nucleotide-disulfide |
| M. thermoresistible (build 8) | TH_1173 | ndh | 1e-98 | 45.43% (416) | PROBABLE NADH DEHYDROGENASE NDH |
| M. ulcerans Agy99 | MUL_3025 | ndh | 1e-102 | 47.23% (415) | NADH dehydrogenase Ndh |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1885c|M.bovis_AF2122/97 ---------------MSPQQEPTAQPPRRHRVVIIGSGFGGLNAAKKLKR
Rv1854c|M.tuberculosis_H37Rv ---------------MSPQQEPTAQPPRRHRVVIIGSGFGGLNAAKKLKR
MLBr_02061|M.leprae_Br4923 ---------------MNAQPAATAAQDRRHQVVIIGSGFGGLNAAKKLKR
MMAR_2728|M.marinum_M ---------------MSPQPETTAQARPRHRVVIIGSGFGGLNAAKKLKR
MUL_3025|M.ulcerans_Agy99 ---------------MSPQPETTAQARPRHRVVIIGSGFGGLNAAKKLKR
MAV_2867|M.avium_104 ---------------MSPHSGSTAGPERRHQVVIIGSGFGGLNAAKKLKH
TH_1173|M.thermoresistible__bu ---------------MS----------DRHKVVIIGSGFGGLNAAKALRR
MAB_2429c|M.abscessus_ATCC_199 MSTTPDAAGAAAPVAANPQPKIQDALTPKHRVVIIGSGFGGLTAAKTLKR
Mvan_2953|M.vanbaalenii_PYR-1 ------------------------MVGRRLRVVIIGGGFGGLFCARRLAR
MSMEG_3457|M.smegmatis_MC2_155 -----------------------------MKVLVIGGGFGGLFCARRLGR
Mflv_3459|M.gilvum_PYR-GCK ------------------------MSAARPRVVIIGGGFGGLFCARRLAR
:*::**.***** .*: * :
Mb1885c|M.bovis_AF2122/97 ADVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQRNVQVL
Rv1854c|M.tuberculosis_H37Rv ADVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQRNVQVL
MLBr_02061|M.leprae_Br4923 ANVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQRNIQVL
MMAR_2728|M.marinum_M ADVDIKLIARTTHHLFQPLLYQVATGIISEGDIAPPTRVVLRRQRNAQVL
MUL_3025|M.ulcerans_Agy99 ADVDIKLIARTTHHLFQPLLYQVATGIISEGDIAPPTRVVLRRQRNAQVL
MAV_2867|M.avium_104 ANVDIKLIARTTHHLFQPLLYQVATGIVSEGDIAPPTRVVLRRQRNVQVL
TH_1173|M.thermoresistible__bu ADVDVKLIARTTHHLFQPLLYQVATGIISEGEIAPATRVVLRRQRNVQVL
MAB_2429c|M.abscessus_ATCC_199 ANADVTLIARTTHHLFQPLLYQVATGIISEGEIAPPTRQILKDQDNAQVV
Mvan_2953|M.vanbaalenii_PYR-1 ADVDVTVLDRTAGHLFQPLLYQCATGTLSIGHISRPLREEFAGHPNITTL
MSMEG_3457|M.smegmatis_MC2_155 LDVDVTLVDRAAGHLFQPLLYQCATGTLSIGHISRPLREELARYSNVKPL
Mflv_3459|M.gilvum_PYR-GCK VDVDVTLLDRAACHVFQPLLYQCATGTLSIGQISRSLREELEHHENVRTL
:.*:.:: *:: *:******* *** :* *.*: . * : * :
Mb1885c|M.bovis_AF2122/97 LGNVTHIDLAGQCVVSELLG-HTYQTPYDSLIVAAGAGQSYFGNDHFAEF
Rv1854c|M.tuberculosis_H37Rv LGNVTHIDLAGQCVVSELLG-HTYQTPYDSLIVAAGAGQSYFGNDHFAEF
MLBr_02061|M.leprae_Br4923 LGNVTHIDLANQCVVSELLG-HTYETLYDSLIVAAGAGQSYFGNDQFAEF
MMAR_2728|M.marinum_M LGNVTHIDLAKQSVVSDLLG-HTYETPYDTLIVAAGAGQSYFGNDHFAEF
MUL_3025|M.ulcerans_Agy99 LGNVTHIDLAKQSVVSDLLG-HTYETPYDTLIVAAGAGQSYFGNDHFAEF
MAV_2867|M.avium_104 LGDVTHIDLAGKFVVSDLLG-HTYETPYDTLIVAAGAGQSYFGNDHFAEF
TH_1173|M.thermoresistible__bu LGDVTRIDLTNRRVESVLLG-HTYSTPYDSLIVAAGAGQSYFGNDHFAEF
MAB_2429c|M.abscessus_ATCC_199 LGDVTSIDLEKQVVHSSLLG-HDYSTPYDSLIVAAGAGQSYFGNDHFAEW
Mvan_2953|M.vanbaalenii_PYR-1 LGEAVALDPERRTVTALRPDETTFTIDYDVLVVAAGMQQSYFGKPHFAQW
MSMEG_3457|M.smegmatis_MC2_155 LGEAIRLDPDQRQVTFLRPDETSFVLDYDVLVVAAGMRQSYMGNERFSEW
Mflv_3459|M.gilvum_PYR-GCK LGEAIRLDPAGRRVTARRPDDSTFDLDYDYLVIAAGMRQSYFGNEDFAAW
**:. :* : * . : ** *::*** ***:*: *: :
Mb1885c|M.bovis_AF2122/97 APGMKSIDDALELRGRILSAFEQAERSSDPERRAKLLTFTVVGAGPTGVE
Rv1854c|M.tuberculosis_H37Rv APGMKSIDDALELRGRILSAFEQAERSSDPERRAKLLTFTVVGAGPTGVE
MLBr_02061|M.leprae_Br4923 APGMKSIDDALELRGRILSAFEQAERSNDPERREKLLTFTVVGAGPTGVE
MMAR_2728|M.marinum_M APGMKSIDDALELRGRILSAFEQAERSRDAERRAKLLTFTVVGAGPTGVE
MUL_3025|M.ulcerans_Agy99 APGMKSIDDALELRGRILSAFEQAERSRDAERRAKLLTFTVVGAGPTGVE
MAV_2867|M.avium_104 APGMKSIDDALEVRGRILSAFEQAERSRDPERRAKLLTFTVIGAGPTGVE
TH_1173|M.thermoresistible__bu APGMKTIDDALELRARILGAFEQAERSSDAKRREKLLTFTVVGAGPTGVE
MAB_2429c|M.abscessus_ATCC_199 APGMKSIDDALELRGRILGAFEQAERSSDPVRRRKLMTFTVVGAGPTGVE
Mvan_2953|M.vanbaalenii_PYR-1 APGMKTLDDALSIRQRIFTAFEIAETLPPGAERDAWLTFAVTGGGPTGVE
MSMEG_3457|M.smegmatis_MC2_155 APGMKTLDDALSIRQRVFTAFEIAETLPPGPERDAWLTFAVAGGGPTGVE
Mflv_3459|M.gilvum_PYR-GCK APGMKTLDDALSIRRRVFAAFEIAETLPPGPERDQWLTFAVTGAGPTGVE
*****::****.:* *:: *** ** .* :**:* *.******
Mb1885c|M.bovis_AF2122/97 MAGQIAELAEHTLKGAFRHID-STKARVILLDAAPAVLPPMGAKLGQRAA
Rv1854c|M.tuberculosis_H37Rv MAGQIAELAEHTLKGAFRHID-STKARVILLDAAPAVLPPMGAKLGQRAA
MLBr_02061|M.leprae_Br4923 MAGQIAELADHTLKGAFRRID-STKARVVLLDAAPAVLPPMGEKLGQRAA
MMAR_2728|M.marinum_M MAGQIAELAEHTLKGAFRHID-STRARVILLDAAPAVLPPFGDKLGERAA
MUL_3025|M.ulcerans_Agy99 MAGQIAELAEHTLKGAFRHID-STRARVILLDAAPAVLPPFGDKLGERAA
MAV_2867|M.avium_104 MAGQIAELATYTLKGSFRHID-PTKARVILLDAAPAVLPPFGDKLGKRAA
TH_1173|M.thermoresistible__bu MAGQIAELAKYTLKGTFRSIDPATEARVILLDAAPAVLPTMGEKLGKKAQ
MAB_2429c|M.abscessus_ATCC_199 MAGQIAELANETLKGTFRHID-PTDARVILLDAAPAVLPPFGPKLGDKAR
Mvan_2953|M.vanbaalenii_PYR-1 LAGQIREMATRTLTNEFHSIE-PEEARVLLFDGGDRVLKSFAPELSGRAA
MSMEG_3457|M.smegmatis_MC2_155 LAGQIREMATRTLANEFHSIE-PEEARVLLFDGGDRVLKSFAPDLAERAA
Mflv_3459|M.gilvum_PYR-GCK LAGQIRELATRALANQFHSIE-PEDARVLLFDGGDRVLKTFAPDLSARAQ
:**** *:* :* . *: *: . ***:*:*.. ** .:. .*. :*
Mb1885c|M.bovis_AF2122/97 ARLQKLGVEIQLGAMVTDVDRNGITVKDSDG-TVRRIESACKVWSAGVSA
Rv1854c|M.tuberculosis_H37Rv ARLQKLGVEIQLGAMVTDVDRNGITVKDSDG-TVRRIESACKVWSAGVSA
MLBr_02061|M.leprae_Br4923 ARLQKMGVEIQLSAMVTDVDRNGITVQDSDG-TVRRIESACKVWSAGVSA
MMAR_2728|M.marinum_M ARLQKLGVEIQLGAMVTDVDRNGITVKDSDG-TIRRIESACKVWSAGVQA
MUL_3025|M.ulcerans_Agy99 ARLQKLGVEIQLGAMVTDVDRNGITVKDSDG-TIRRIESACKVWSAGVQA
MAV_2867|M.avium_104 DRLEKMGVEIQLGAMVTDVDRNGITVKDSDG-TVRRIESACKVWSAGVSA
TH_1173|M.thermoresistible__bu ARLEKMGVEIQLGAMVTDVDRNGITVKDSDG-TVRRIESACKVWSAGVSA
MAB_2429c|M.abscessus_ATCC_199 KRLEKLGVEIQLSAMVTDVDRNGLTVKHADG-SIERIESWCKVWSAGVSA
Mvan_2953|M.vanbaalenii_PYR-1 GTLRSLGVDVRLGVHVTDVQREGVTVAPKSGGSEEYYAVRTVLWTAGVEA
MSMEG_3457|M.smegmatis_MC2_155 ESLSALGVELRMGVHVTDVRRDGVTVTPKNGGPAEEYATRTVLWTAGVEA
Mflv_3459|M.gilvum_PYR-GCK RVLEDLGVEMRFGVRVSDVRRDGITVSPKDGSPDADVATRTVLWTAGVEA
* :**::::.. *:** *:*:** .* . :*:***.*
Mb1885c|M.bovis_AF2122/97 SWLGRDLAEQSRVELDRAGRVQVLPDLSIPGYPNVFVVGDMAAVEGVPGV
Rv1854c|M.tuberculosis_H37Rv SRLGRDLAEQSRVELDRAGRVQVLPDLSIPGYPNVFVVGDMAAVEGVPGV
MLBr_02061|M.leprae_Br4923 SRLGRDLAEQSLVELDWAGRVKVLPDLSIPGYRNVFVVGDMAAIDGVPGV
MMAR_2728|M.marinum_M SRLGRDLAEQSEVELDRAGRVQVLPDLSVPGHPNVFVIGDMAAVEGVPGV
MUL_3025|M.ulcerans_Agy99 SRLGRDLAEQSEVELDRAGRVQVLPDLSVPGHPNVFVIGDMAAVEGVPGV
MAV_2867|M.avium_104 SPLGRDLAEQSTVELDRAGRVKVLPDLSIPGHPNVFVIGDLAAVEGVPGV
TH_1173|M.thermoresistible__bu SPLGKDLAEQSGAELDRAGRVKVLPDLSLPGHPNVFVVGDMAAVEGVPGM
MAB_2429c|M.abscessus_ATCC_199 SPLGKNLAEQSGVELDRAGRVKVGPDLSIPGHPNVFVVGDMMAVDGVPGV
Mvan_2953|M.vanbaalenii_PYR-1 VPFAGHLAEVLDVQADRAGRLAVQPDLSVPGHPEIFVIGDLVGRDDLPGV
MSMEG_3457|M.smegmatis_MC2_155 VPFARHVAEVLGAKTDRAGRIEVEPDLTVPGHPEVFVVGDLVGRDKLPGV
Mflv_3459|M.gilvum_PYR-GCK VPFARHAAEVLGATTDRSGRIAVDADLSVPNHREVFVIGDLAGRDGLPGV
:. . ** . * :**: * .**::*.: ::**:**: . : :**:
Mb1885c|M.bovis_AF2122/97 AQGAIQGAKYVASTIKAELAGANPAEREPFQYFDKGSMATVSRFSAVAKI
Rv1854c|M.tuberculosis_H37Rv AQGAIQGAKYVASTIKAELAGANPAEREPFQYFDKGSMATVSRFSAVAKI
MLBr_02061|M.leprae_Br4923 AQGAIQGAKYVANNIKAELGEANPAEREPFQYFDKGSMATVSRFSAVAKI
MMAR_2728|M.marinum_M AQGAIQGAKYVATTIKSELAGANAAEREPFQYFDKGSMATVSRFSAVAKI
MUL_3025|M.ulcerans_Agy99 AQGAIQGAKYVATTIKSELAGANAAEREPFQYFDKGSMATVSRFSAVAKI
MAV_2867|M.avium_104 AQGAIQGAKYVANTIKAELGGADPAEREPFQYFDKGSMATVSRFSAVAKI
TH_1173|M.thermoresistible__bu AQGAIQGARHAAKLIKNELKGADPADREPFQYFDKGSMATVSRFSAVAKI
MAB_2429c|M.abscessus_ATCC_199 AQGAIQGGRYAAKQIKAELKGRSPAERAPFKFFDKGSMATVSRYSAVVKI
Mvan_2953|M.vanbaalenii_PYR-1 AENAMQGGLHVAACIRRDLAGR---PRRNYRYRDLGSAAYISRGNALLQV
MSMEG_3457|M.smegmatis_MC2_155 AENAMQGGLHVARCVRRDLAGK---PRRPYRYHDLGSAAYIRRGDALLQV
Mflv_3459|M.gilvum_PYR-GCK AENAMQGGMHAAACMRRDLQGK---RRKNFRYRDLGSAAYISRGHALMQL
*:.*:**. :.* :: :* * ::: * ** * : * *: ::
Mb1885c|M.bovis_AF2122/97 GPVEFSGFIAWLIWLVLHLAYLIGFKTKITTLLSWTVTFLSTRR------
Rv1854c|M.tuberculosis_H37Rv GPVEFSGFIAWLIWLVLHLAYLIGFKTKITTLLSWTVTFLSTRR------
MLBr_02061|M.leprae_Br4923 GPLEFSGFIAWLMWLVLHLMYLIGFKTKITTLLSWTVTFLSTRR------
MMAR_2728|M.marinum_M GPVEISGFIAWLAWLVLHLVYLVGFKTKVSTLLSWTVTFLSTRR------
MUL_3025|M.ulcerans_Agy99 GPVEISGFIAWLAWLVLHLVYLVGFKTKVSTLLSWTVTFLSTRR------
MAV_2867|M.avium_104 GPLEFSGLFAWFAWLVLHLVYLVGFKTKVSTLLSWTVTFLSTRR------
TH_1173|M.thermoresistible__bu GPLEFGGFIAWLAWLFLHLVYIVGFKARLTTALSWIVTFLGSHR------
MAB_2429c|M.abscessus_ATCC_199 GKIEFSGFIAWLSWLALHLVYLVGFKNRLVTLILWSIAFVTRSR------
Mvan_2953|M.vanbaalenii_PYR-1 GPVKVAGFLGWIAWGLLHIAFLTGVRNRVSTVVTWLATIARASRYHRAFM
MSMEG_3457|M.smegmatis_MC2_155 HRVRVSGFAGWLAWGFIHIAFLTGMRNRVSTVITWMATIARASRYHRAFM
Mflv_3459|M.gilvum_PYR-GCK GPVKLSGLSGWIGWGFIHIAFLTGVRNRFSTVGTWLATIARSNRSDRTFI
:...*: .*: * :*: :: *.: :. * * :: *
Mb1885c|M.bovis_AF2122/97 -GQLTITDQQAFARTRLEQLAELAAEAQGSAASAKVAS------------
Rv1854c|M.tuberculosis_H37Rv -GQLTITDQQAFARTRLEQLAELAAEAQGSAASAKVAS------------
MLBr_02061|M.leprae_Br4923 -GQLTITEQQAFARTRLEQLAELTAESQSTAAANRATMRAS---------
MMAR_2728|M.marinum_M -GQLTITEQQAFARTRLEQLAELASEAQSTEARQAS--------------
MUL_3025|M.ulcerans_Agy99 -GQLTITEQQAFARTRLEQLAELASEAQSTEARQAS--------------
MAV_2867|M.avium_104 -GQLTITEQQAFARTRLEQLAVLAAETKRPAARRAS--------------
TH_1173|M.thermoresistible__bu -GQLTITEQQAFARTRIEELQEIAASVQDTQKAAS---------------
MAB_2429c|M.abscessus_ATCC_199 -GQMAITEQQAWARTRLDQLEELVAEKNGDEEPTTQPDKEKAAS------
Mvan_2953|M.vanbaalenii_PYR-1 LGS-VSAPEARYTWSSRDQPTPPQALCEDPRTGHTSHMTISAQDVRRLLD
MSMEG_3457|M.smegmatis_MC2_155 LGDPTKIEQERYTWSRWDQESPPPETWPPPAVEQGP--------------
Mflv_3459|M.gilvum_PYR-GCK LGG-SGHPEEPYTWESPVHQG--GSSRDNSGEDKTA--------------
* : :: .
Mb1885c|M.bovis_AF2122/97 --------------------------------------------------
Rv1854c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02061|M.leprae_Br4923 --------------------------------------------------
MMAR_2728|M.marinum_M --------------------------------------------------
MUL_3025|M.ulcerans_Agy99 --------------------------------------------------
MAV_2867|M.avium_104 --------------------------------------------------
TH_1173|M.thermoresistible__bu --------------------------------------------------
MAB_2429c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_2953|M.vanbaalenii_PYR-1 AEHDSTLVLIEGRTEVITGQQKSSDQYRGAFEVISRDELLSRTGGAAHLT
MSMEG_3457|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3459|M.gilvum_PYR-GCK --------------------------------------------------
Mb1885c|M.bovis_AF2122/97 --------------------
Rv1854c|M.tuberculosis_H37Rv --------------------
MLBr_02061|M.leprae_Br4923 --------------------
MMAR_2728|M.marinum_M --------------------
MUL_3025|M.ulcerans_Agy99 --------------------
MAV_2867|M.avium_104 --------------------
TH_1173|M.thermoresistible__bu --------------------
MAB_2429c|M.abscessus_ATCC_199 --------------------
Mvan_2953|M.vanbaalenii_PYR-1 DDQISAQAAALDTAVNELGG
MSMEG_3457|M.smegmatis_MC2_155 --------------------
Mflv_3459|M.gilvum_PYR-GCK --------------------