For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSAARPRVVIIGGGFGGLFCARRLARVDVDVTLLDRAACHVFQPLLYQCATGTLSIGQISRSLREELEHH ENVRTLLGEAIRLDPAGRRVTARRPDDSTFDLDYDYLVIAAGMRQSYFGNEDFAAWAPGMKTLDDALSIR RRVFAAFEIAETLPPGPERDQWLTFAVTGAGPTGVELAGQIRELATRALANQFHSIEPEDARVLLFDGGD RVLKTFAPDLSARAQRVLEDLGVEMRFGVRVSDVRRDGITVSPKDGSPDADVATRTVLWTAGVEAVPFAR HAAEVLGATTDRSGRIAVDADLSVPNHREVFVIGDLAGRDGLPGVAENAMQGGMHAAACMRRDLQGKRRK NFRYRDLGSAAYISRGHALMQLGPVKLSGLSGWIGWGFIHIAFLTGVRNRFSTVGTWLATIARSNRSDRT FILGGSGHPEEPYTWESPVHQGGSSRDNSGEDKTA
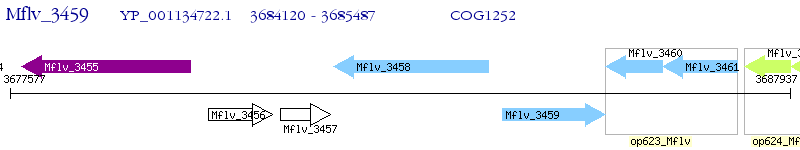
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3459 | - | - | 100% (455) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. gilvum PYR-GCK | Mflv_3348 | - | 1e-92 | 42.24% (419) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1066 | - | 2e-12 | 26.48% (287) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0398c | ndhA | 2e-94 | 45.59% (408) | membrane NADH dehydrogenase |
| M. tuberculosis H37Rv | Rv0392c | ndhA | 9e-95 | 45.83% (408) | membrane NADH dehydrogenase |
| M. leprae Br4923 | MLBr_02061 | - | 3e-89 | 42.24% (419) | putative dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2429c | - | 8e-97 | 44.66% (412) | NADH dehydrogenase (NDH) |
| M. marinum M | MMAR_0689 | ndh_1 | 4e-96 | 45.91% (416) | NADH dehydrogenase Ndh_1 |
| M. avium 104 | MAV_2867 | - | 7e-96 | 45.11% (419) | DoxD family protein/pyridine nucleotide-disulfide |
| M. smegmatis MC2 155 | MSMEG_3457 | - | 0.0 | 74.19% (430) | DoxD family protein/pyridine nucleotide-disulfide |
| M. thermoresistible (build 8) | TH_1173 | ndh | 1e-100 | 45.95% (420) | PROBABLE NADH DEHYDROGENASE NDH |
| M. ulcerans Agy99 | MUL_3025 | ndh | 2e-94 | 44.63% (419) | NADH dehydrogenase Ndh |
| M. vanbaalenii PYR-1 | Mvan_2953 | - | 0.0 | 73.44% (433) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2867|M.avium_104 ---------------MSPHSGSTAGPERRHQVVIIGSGFGGLNAAKKLKH
MUL_3025|M.ulcerans_Agy99 ---------------MSPQPETTAQARPRHRVVIIGSGFGGLNAAKKLKR
MLBr_02061|M.leprae_Br4923 ---------------MNAQPAATAAQDRRHQVVIIGSGFGGLNAAKKLKR
TH_1173|M.thermoresistible__bu ---------------MS----------DRHKVVIIGSGFGGLNAAKALRR
MAB_2429c|M.abscessus_ATCC_199 MSTTPDAAGAAAPVAANPQPKIQDALTPKHRVVIIGSGFGGLTAAKTLKR
Mb0398c|M.bovis_AF2122/97 ---------------MTLSSGEPSAVGGRHRVVIIGSGFGGLNAAKALKR
Rv0392c|M.tuberculosis_H37Rv ---------------MTLSSGEPSAVGGRHRVVIIGSGFGGLNAAKALKR
MMAR_0689|M.marinum_M ---------------MTPPSSESSAAGRRHHVVIIGSGFGGLSAARALKR
MSMEG_3457|M.smegmatis_MC2_155 -----------------------------MKVLVIGGGFGGLFCARRLGR
Mvan_2953|M.vanbaalenii_PYR-1 ------------------------MVGRRLRVVIIGGGFGGLFCARRLAR
Mflv_3459|M.gilvum_PYR-GCK ------------------------MSAARPRVVIIGGGFGGLFCARRLAR
:*::**.***** .*: * :
MAV_2867|M.avium_104 ANVDIKLIARTTHHLFQPLLYQVATGIVSEGDIAPPTRVVLRRQRNVQVL
MUL_3025|M.ulcerans_Agy99 ADVDIKLIARTTHHLFQPLLYQVATGIISEGDIAPPTRVVLRRQRNAQVL
MLBr_02061|M.leprae_Br4923 ANVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQRNIQVL
TH_1173|M.thermoresistible__bu ADVDVKLIARTTHHLFQPLLYQVATGIISEGEIAPATRVVLRRQRNVQVL
MAB_2429c|M.abscessus_ATCC_199 ANADVTLIARTTHHLFQPLLYQVATGIISEGEIAPPTRQILKDQDNAQVV
Mb0398c|M.bovis_AF2122/97 ADVDITLISKTTTHLFQPLLYQVATGILSEGDIAPTTRLILRRQKNVRVL
Rv0392c|M.tuberculosis_H37Rv ADVDITLISKTTTHLFQPLLYQVATGILSEGDIAPTTRLILRRQKNVRVL
MMAR_0689|M.marinum_M ADVDITLISKTTTHLFQPLLYQVATGILSEGDIAPTTRLILRRQKNVRVL
MSMEG_3457|M.smegmatis_MC2_155 LDVDVTLVDRAAGHLFQPLLYQCATGTLSIGHISRPLREELARYSNVKPL
Mvan_2953|M.vanbaalenii_PYR-1 ADVDVTVLDRTAGHLFQPLLYQCATGTLSIGHISRPLREEFAGHPNITTL
Mflv_3459|M.gilvum_PYR-GCK VDVDVTLLDRAACHVFQPLLYQCATGTLSIGQISRSLREELEHHENVRTL
:.*:.:: ::: *:******* *** :* *.*: . * : * :
MAV_2867|M.avium_104 LGDVTHIDLAGKFVVSDLLG-HTYETPYDTLIVAAGAGQSYFGNDHFAEF
MUL_3025|M.ulcerans_Agy99 LGNVTHIDLAKQSVVSDLLG-HTYETPYDTLIVAAGAGQSYFGNDHFAEF
MLBr_02061|M.leprae_Br4923 LGNVTHIDLANQCVVSELLG-HTYETLYDSLIVAAGAGQSYFGNDQFAEF
TH_1173|M.thermoresistible__bu LGDVTRIDLTNRRVESVLLG-HTYSTPYDSLIVAAGAGQSYFGNDHFAEF
MAB_2429c|M.abscessus_ATCC_199 LGDVTSIDLEKQVVHSSLLG-HDYSTPYDSLIVAAGAGQSYFGNDHFAEW
Mb0398c|M.bovis_AF2122/97 LGEVNAIDLKAQTVTSKLMD-MTTVTPYDSLIVAAGAQQSYFGNDEFATF
Rv0392c|M.tuberculosis_H37Rv LGEVNAIDLKAQTVTSKLMD-MTTVTPYDSLIVAAGAQQSYFGNDEFATF
MMAR_0689|M.marinum_M LGEVMGIDLNAQTVTSKLMD-MTTVSPYDSLIVAAGAQQSYFGNDDFAIF
MSMEG_3457|M.smegmatis_MC2_155 LGEAIRLDPDQRQVTFLRPDETSFVLDYDVLVVAAGMRQSYMGNERFSEW
Mvan_2953|M.vanbaalenii_PYR-1 LGEAVALDPERRTVTALRPDETTFTIDYDVLVVAAGMQQSYFGKPHFAQW
Mflv_3459|M.gilvum_PYR-GCK LGEAIRLDPAGRRVTARRPDDSTFDLDYDYLVIAAGMRQSYFGNEDFAAW
**:. :* : * . ** *::*** ***:*: *: :
MAV_2867|M.avium_104 APGMKSIDDALEVRGRILSAFEQAERSRDPERRAKLLTFTVIGAGPTGVE
MUL_3025|M.ulcerans_Agy99 APGMKSIDDALELRGRILSAFEQAERSRDAERRAKLLTFTVVGAGPTGVE
MLBr_02061|M.leprae_Br4923 APGMKSIDDALELRGRILSAFEQAERSNDPERREKLLTFTVVGAGPTGVE
TH_1173|M.thermoresistible__bu APGMKTIDDALELRARILGAFEQAERSSDAKRREKLLTFTVVGAGPTGVE
MAB_2429c|M.abscessus_ATCC_199 APGMKSIDDALELRGRILGAFEQAERSSDPVRRRKLMTFTVVGAGPTGVE
Mb0398c|M.bovis_AF2122/97 APGMKTIDDALELRGRILGAFEAAEVSTDHAERERRLTFVVVGAGPTGVE
Rv0392c|M.tuberculosis_H37Rv APGMKTIDDALELRGRILGAFEAAEVSTDHAERERRLTFVVVGAGPTGVE
MMAR_0689|M.marinum_M APGMKTIDDALELRGRILGAFEAAEVATDHDERERRLTFVVVGAGPTGVE
MSMEG_3457|M.smegmatis_MC2_155 APGMKTLDDALSIRQRVFTAFEIAETLPPGPERDAWLTFAVAGGGPTGVE
Mvan_2953|M.vanbaalenii_PYR-1 APGMKTLDDALSIRQRIFTAFEIAETLPPGAERDAWLTFAVTGGGPTGVE
Mflv_3459|M.gilvum_PYR-GCK APGMKTLDDALSIRRRVFAAFEIAETLPPGPERDQWLTFAVTGAGPTGVE
*****::****.:* *:: *** ** .* :**.* *.******
MAV_2867|M.avium_104 MAGQIAELATYTLKGSFRHID-PTKARVILLDAAPAVLPPFGDKLGKRAA
MUL_3025|M.ulcerans_Agy99 MAGQIAELAEHTLKGAFRHID-STRARVILLDAAPAVLPPFGDKLGERAA
MLBr_02061|M.leprae_Br4923 MAGQIAELADHTLKGAFRRID-STKARVVLLDAAPAVLPPMGEKLGQRAA
TH_1173|M.thermoresistible__bu MAGQIAELAKYTLKGTFRSIDPATEARVILLDAAPAVLPTMGEKLGKKAQ
MAB_2429c|M.abscessus_ATCC_199 MAGQIAELANETLKGTFRHID-PTDARVILLDAAPAVLPPFGPKLGDKAR
Mb0398c|M.bovis_AF2122/97 VAGQIVELAERTLAGAFRTIT-PSECRVILLDAAPAVLPPMGPKLGLKAQ
Rv0392c|M.tuberculosis_H37Rv VAGQIVELAERTLAGAFRTIT-PSECRVILLDAAPAVLPPMGPKLGLKAQ
MMAR_0689|M.marinum_M LAGEIVQLAERTLAGAFRTIT-PSECRVILLDAAPAVLPPMGPKLGLKAQ
MSMEG_3457|M.smegmatis_MC2_155 LAGQIREMATRTLANEFHSIE-PEEARVLLFDGGDRVLKSFAPDLAERAA
Mvan_2953|M.vanbaalenii_PYR-1 LAGQIREMATRTLTNEFHSIE-PEEARVLLFDGGDRVLKSFAPELSGRAA
Mflv_3459|M.gilvum_PYR-GCK LAGQIRELATRALANQFHSIE-PEDARVLLFDGGDRVLKTFAPDLSARAQ
:**:* ::* :* . *: * . .**:*:*.. ** .:. .*. :*
MAV_2867|M.avium_104 DRLEKMGVEIQLGAMVTDVDRNGITVKDSDGT-VRRIESACKVWSAGVSA
MUL_3025|M.ulcerans_Agy99 ARLQKLGVEIQLGAMVTDVDRNGITVKDSDGT-IRRIESACKVWSAGVQA
MLBr_02061|M.leprae_Br4923 ARLQKMGVEIQLSAMVTDVDRNGITVQDSDGT-VRRIESACKVWSAGVSA
TH_1173|M.thermoresistible__bu ARLEKMGVEIQLGAMVTDVDRNGITVKDSDGT-VRRIESACKVWSAGVSA
MAB_2429c|M.abscessus_ATCC_199 KRLEKLGVEIQLSAMVTDVDRNGLTVKHADGS-IERIESWCKVWSAGVSA
Mb0398c|M.bovis_AF2122/97 RRLEKMDAEVQLNAMVTAVDYKGITIKEKDGG-ERRIECACKVWAAGVAA
Rv0392c|M.tuberculosis_H37Rv RRLEKMDVEVQLNAMVTAVDYKGITIKEKDGG-ERRIECACKVWAAGVAA
MMAR_0689|M.marinum_M KKLEKMDVEIQLNAMVTAVDYKGITVKDKDGT-ERRIDCACKVWAAGVQA
MSMEG_3457|M.smegmatis_MC2_155 ESLSALGVELRMGVHVTDVRRDGVTVTPKNGGPAEEYATRTVLWTAGVEA
Mvan_2953|M.vanbaalenii_PYR-1 GTLRSLGVDVRLGVHVTDVQREGVTVAPKSGGSEEYYAVRTVLWTAGVEA
Mflv_3459|M.gilvum_PYR-GCK RVLEDLGVEMRFGVRVSDVRRDGITVSPKDGSPDADVATRTVLWTAGVEA
* :..::::.. *: * .*:*: .* :*:*** *
MAV_2867|M.avium_104 SPLGRDLAEQS-TVELDRAGRVKVLPDLSIPGHPNVFVIGDLAAVEGVPG
MUL_3025|M.ulcerans_Agy99 SRLGRDLAEQS-EVELDRAGRVQVLPDLSVPGHPNVFVIGDMAAVEGVPG
MLBr_02061|M.leprae_Br4923 SRLGRDLAEQS-LVELDWAGRVKVLPDLSIPGYRNVFVVGDMAAIDGVPG
TH_1173|M.thermoresistible__bu SPLGKDLAEQS-GAELDRAGRVKVLPDLSLPGHPNVFVVGDMAAVEGVPG
MAB_2429c|M.abscessus_ATCC_199 SPLGKNLAEQS-GVELDRAGRVKVGPDLSIPGHPNVFVVGDMMAVDGVPG
Mb0398c|M.bovis_AF2122/97 SPLGKMIAEGSDGTEIDRAGRVIVEPDLTVKGHPNVFVVGDLMFVPGVPG
Rv0392c|M.tuberculosis_H37Rv SPLGKMIAEGSDGTEIDRAGRVIVEPDLTVKGHPNVFVVGDLMFVPGVPG
MMAR_0689|M.marinum_M SPLGKMIAEQSDGTETDRAGRVIVEPDLTVKGHPHVFVIGDLMSVPGVPG
MSMEG_3457|M.smegmatis_MC2_155 VPFARHVAEVL-GAKTDRAGRIEVEPDLTVPGHPEVFVVGDLVGRDKLPG
Mvan_2953|M.vanbaalenii_PYR-1 VPFAGHLAEVL-DVQADRAGRLAVQPDLSVPGHPEIFVIGDLVGRDDLPG
Mflv_3459|M.gilvum_PYR-GCK VPFARHAAEVL-GATTDRSGRIAVDADLSVPNHREVFVIGDLAGRDGLPG
:. ** . * :**: * .**:: .: .:**:**: :**
MAV_2867|M.avium_104 VAQGAIQGAKYVANTIKAELGG-ADPAEREPFQYFDKGSMATVSRFSAVA
MUL_3025|M.ulcerans_Agy99 VAQGAIQGAKYVATTIKSELAG-ANAAEREPFQYFDKGSMATVSRFSAVA
MLBr_02061|M.leprae_Br4923 VAQGAIQGAKYVANNIKAELGE-ANPAEREPFQYFDKGSMATVSRFSAVA
TH_1173|M.thermoresistible__bu MAQGAIQGARHAAKLIKNELKG-ADPADREPFQYFDKGSMATVSRFSAVA
MAB_2429c|M.abscessus_ATCC_199 VAQGAIQGGRYAAKQIKAELKG-RSPAERAPFKFFDKGSMATVSRYSAVV
Mb0398c|M.bovis_AF2122/97 VAQGAIQGARYATTVIKHMVKGNDDPANRKPFHYFNKGSMATISRHSAVA
Rv0392c|M.tuberculosis_H37Rv VAQGAIQGARYATTVIKHMVKGNDDPANRKPFHYFNKGSMATISRHSAVA
MMAR_0689|M.marinum_M MAQGAIQGAHYATKTIKQAVKGHDDPANRKPFQYFNKGSMALISHYSAVA
MSMEG_3457|M.smegmatis_MC2_155 VAENAMQGGLHVARCVRRDLAG----KPRRPYRYHDLGSAAYIRRGDALL
Mvan_2953|M.vanbaalenii_PYR-1 VAENAMQGGLHVAACIRRDLAG----RPRRNYRYRDLGSAAYISRGNALL
Mflv_3459|M.gilvum_PYR-GCK VAENAMQGGMHAAACMRRDLQG----KRRKNFRYRDLGSAAYISRGHALM
:*:.*:**. :.: :: : * ::: : ** * : : *:
MAV_2867|M.avium_104 KIGPLEFSGLFAWFAWLVLHLVYLVGFKTKVSTLLSWTVTFLSTRRGQLT
MUL_3025|M.ulcerans_Agy99 KIGPVEISGFIAWLAWLVLHLVYLVGFKTKVSTLLSWTVTFLSTRRGQLT
MLBr_02061|M.leprae_Br4923 KIGPLEFSGFIAWLMWLVLHLMYLIGFKTKITTLLSWTVTFLSTRRGQLT
TH_1173|M.thermoresistible__bu KIGPLEFGGFIAWLAWLFLHLVYIVGFKARLTTALSWIVTFLGSHRGQLT
MAB_2429c|M.abscessus_ATCC_199 KIGKIEFSGFIAWLSWLALHLVYLVGFKNRLVTLILWSIAFVTRSRGQMA
Mb0398c|M.bovis_AF2122/97 QVGKLEFAGYFAWLAWLVLHLVYLVGYRNRIAALFAWGISFMGRARGQMA
Rv0392c|M.tuberculosis_H37Rv QVGKLEFAGYFAWLAWLVLHLVYLVGYRNRIAALFAWGISFMGRARGQMA
MMAR_0689|M.marinum_M KVGKVEFAGFIAWLAWLFLHLLYLVGHRNRVAAAFSWGLAFLGRTRGQMA
MSMEG_3457|M.smegmatis_MC2_155 QVHRVRVSGFAGWLAWGFIHIAFLTGMRNRVSTVITWMATIARASRYHRA
Mvan_2953|M.vanbaalenii_PYR-1 QVGPVKVAGFLGWIAWGLLHIAFLTGVRNRVSTVVTWLATIARASRYHRA
Mflv_3459|M.gilvum_PYR-GCK QLGPVKLSGLSGWIGWGFIHIAFLTGVRNRFSTVGTWLATIARSNRSDRT
:: :...* .*: * :*: :: * : :. : * :: * . :
MAV_2867|M.avium_104 ITEQQAFARTRLEQLAVLAAET---KRPAAR-----RAS-----------
MUL_3025|M.ulcerans_Agy99 ITEQQAFARTRLEQLAELASEA---QSTEAR-----QAS-----------
MLBr_02061|M.leprae_Br4923 ITEQQAFARTRLEQLAELTAES---QSTAAANRATMRAS-----------
TH_1173|M.thermoresistible__bu ITEQQAFARTRIEELQEIAASV---QDTQKA------AS-----------
MAB_2429c|M.abscessus_ATCC_199 ITEQQAWARTRLDQLEELVAEKNGDEEPTTQPDKEKAAS-----------
Mb0398c|M.bovis_AF2122/97 ITSQMIYARLVMTLMEQQAQG-ALAAAEQAEHAEQEAAG-----------
Rv0392c|M.tuberculosis_H37Rv ITSQMIYARLVMTLMEQQAQG-ALAAAEQAEHAEQEAAG-----------
MMAR_0689|M.marinum_M ITSQMVYARLAMRYLQARAEEDALSAAEHAEQAEQRATG-----------
MSMEG_3457|M.smegmatis_MC2_155 FMLGDPTKIEQERYTWSRWDQESPPPETWPPPAVEQGP------------
Mvan_2953|M.vanbaalenii_PYR-1 FMLGS-VSAPEARYTWSSRDQPTPPQALCEDPRTGHTSHMTISAQDVRRL
Mflv_3459|M.gilvum_PYR-GCK FILGG-SGHPEEPYTWESPVHQG--GSSRDNSGEDKTA------------
: .
MAV_2867|M.avium_104 --------------------------------------------------
MUL_3025|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02061|M.leprae_Br4923 --------------------------------------------------
TH_1173|M.thermoresistible__bu --------------------------------------------------
MAB_2429c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0398c|M.bovis_AF2122/97 --------------------------------------------------
Rv0392c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0689|M.marinum_M --------------------------------------------------
MSMEG_3457|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_2953|M.vanbaalenii_PYR-1 LDAEHDSTLVLIEGRTEVITGQQKSSDQYRGAFEVISRDELLSRTGGAAH
Mflv_3459|M.gilvum_PYR-GCK --------------------------------------------------
MAV_2867|M.avium_104 ----------------------
MUL_3025|M.ulcerans_Agy99 ----------------------
MLBr_02061|M.leprae_Br4923 ----------------------
TH_1173|M.thermoresistible__bu ----------------------
MAB_2429c|M.abscessus_ATCC_199 ----------------------
Mb0398c|M.bovis_AF2122/97 ----------------------
Rv0392c|M.tuberculosis_H37Rv ----------------------
MMAR_0689|M.marinum_M ----------------------
MSMEG_3457|M.smegmatis_MC2_155 ----------------------
Mvan_2953|M.vanbaalenii_PYR-1 LTDDQISAQAAALDTAVNELGG
Mflv_3459|M.gilvum_PYR-GCK ----------------------