For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RTLLGRWQTWAGFGAAALVLFVVAPAVLSDFRLSLLGKFLCFAIVAVGIGLAWGRGGMLVLGQGVFFGLG GYMMAMHLKIADAQLRGDEVPDFMQIQGIRELPGYWAPFASPLVTLIAIVVVPTGIAAALGLGVFKRKVK GAYFAILSQALAAALAILLVGQTSLGGSNGLNRFRTFFGFALNDPVNRRMLYFIAAAVLLVVVAVVRQLM QSRYGELLVAVRDGEERVRFLGYDPANIKVVAYAAAALFASIAGALFAPIVGFIAPSQVGILPSIAFLIG VAIGGRTTLLGPVLGAIGVAWAQTLFSERFPSEWTYAQGLLFIVVVGFFPAGLAGLGVLLKRRRKAGTEP PPEKAPDTDPDTEKVGAGS*
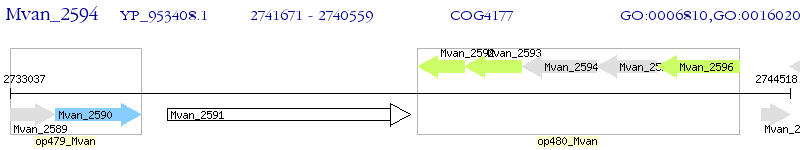
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2594 | - | - | 100% (370) | inner-membrane translocator |
| M. vanbaalenii PYR-1 | Mvan_2843 | - | 3e-18 | 25.09% (283) | inner-membrane translocator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3800 | - | 0.0 | 91.89% (370) | inner-membrane translocator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2624c | - | 7e-16 | 22.97% (283) | high-affinity branched-chain amino acid ABC transporter (LivM) |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2980 | - | 0.0 | 85.60% (375) | hypothetical protein MSMEG_2980 |
| M. thermoresistible (build 8) | TH_2330 | - | 1e-175 | 88.22% (348) | putative membrane protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2594|M.vanbaalenii_PYR-1 -------MR-TLLGRWQTWAG------FGAAALVLFVVAPAVLSDFRLS-
Mflv_3800|M.gilvum_PYR-GCK -------MR-ALLGRWQTWAG------FGVAALVLFVLAPAILSDFRLS-
TH_2330|M.thermoresistible__bu ---VLDRMPGGVLGRWQTWAG------FGLAALVLFVVAPMVLSDFRLG-
MSMEG_2980|M.smegmatis_MC2_155 -------MR-TMLGRWQTWAG------FAVAAVLLFGVAPAVLSDFRLS-
MAB_2624c|M.abscessus_ATCC_199 MSAARALAPGDSIRRWWSELARPVQWVYGVVGFALLALLPVWKIPYLDTP
: ** : . :. ... *: : * :
Mvan_2594|M.vanbaalenii_PYR-1 ------LLGKFLCFAIVAVGIGLAWGRGGMLVLGQGVFFGLGGYMMAMHL
Mflv_3800|M.gilvum_PYR-GCK ------LLGKFLCFAIVAVGIGLAWGRGGMLVLGQGVFFGLGGYMMAMHL
TH_2330|M.thermoresistible__bu ------LFGKFLCFAIVAVGIGLAWGRGGMLVLGQGVFFGIGGYLMGMHL
MSMEG_2980|M.smegmatis_MC2_155 ------LLGKFLCFAIVAVGIGLAWGRGGMLVLGQGVFFGLGGYMMGMHL
MAB_2624c|M.abscessus_ATCC_199 NTSFGGVMAQFAMVALIAIGLNVVVGQAGLLDLGYVGFYAIGAYTVAL-L
::.:* .*::*:*:.:. *:.*:* ** *:.:*.* :.: *
Mvan_2594|M.vanbaalenii_PYR-1 KIADAQLRGDEVPDFMQIQGIRELPGYWAPFASPLVTLIAIVVVPTGIAA
Mflv_3800|M.gilvum_PYR-GCK KIADAQLRGDDVPDFMQIQGIRELPSYWTPFASPAVTLIAIVVIPTGIAV
TH_2330|M.thermoresistible__bu KIADAQIYGTGVPDFMQIAGVRELPGYWEPFASPVVTLAAIVVLPTGIAA
MSMEG_2980|M.smegmatis_MC2_155 KIADAEIRHQAVPDFMQIAGVRELPGYWTPFASPVFTLLAILVIPAGIAA
MAB_2624c|M.abscessus_ATCC_199 TSPDSPWNRMGTSAFFSSD--------WAWLS----CLPIAVAITACFGL
. .*: .. *:. * :: * :.:.: :.
Mvan_2594|M.vanbaalenii_PYR-1 ALGLGVFKRKVKGAYFAILSQALAAALAILLVGQTSLGGSNGLNRFR---
Mflv_3800|M.gilvum_PYR-GCK VLGLGVFKRKVKGAYFAILSQALAAALAILLVGQTSLGGSNGLNRFR---
TH_2330|M.thermoresistible__bu ALGFGVFKRRVKGAYFAILSQALAAALAIFLVGQTSLGGSNGLNRFR---
MSMEG_2980|M.smegmatis_MC2_155 LLGLGVFKRRVKGAYFAILSQALAAALAILLVGQTSLGGSNGLNNFR---
MAB_2624c|M.abscessus_ATCC_199 ILGTPTLR--LRGDYLAIVTLGFGEIIRLLADNLDVTNGPRGLHKVAYPH
** .:: ::* *:**:: .:. : :: . .*..**:..
Mvan_2594|M.vanbaalenii_PYR-1 ----TFFGFALNDPVNRR-------MLYFIAAAVLLVVVAVVRQLMQSRY
Mflv_3800|M.gilvum_PYR-GCK ----TFFGFTLNDPANRR-------MLYFIAAAVLLLVVAVVRQLMQSRY
TH_2330|M.thermoresistible__bu ----TFFGFTLSDPANRR-------MLYFIAAATLLIVVAVARQLMQSRY
MSMEG_2980|M.smegmatis_MC2_155 ----TFFGFTLNDPVNKR-------MLYFIAAATLLIVVAVVRQLMYSRY
MAB_2624c|M.abscessus_ATCC_199 VGEQHVHGGGVFSSANSGGHLNYGVWWYWLGLCLIAAVLVLMGNLERSRV
..* : ...* *::. . : *:.: :* **
Mvan_2594|M.vanbaalenii_PYR-1 GELLVAVRDGEERVRFLGYDPANIKVVAYAAAALFASIAGALFAPIVGFI
Mflv_3800|M.gilvum_PYR-GCK GELLVAVRDGEERVRFLGYDPANIKVVAYAVAALFASIAGALFAPIVGFI
TH_2330|M.thermoresistible__bu GELLVAVRDNEERVRFLGYDPANVKVVAYTVAALFASIAGALFAPIVGFI
MSMEG_2980|M.smegmatis_MC2_155 GELLVAVRDGEERVRFLGYDPANIKVVAYTLAALFASIAGALFTPIVGFI
MAB_2624c|M.abscessus_ATCC_199 GRSWVAIREDEDAAEMMGVPTFRFKLWAFVIGASIGGLSGALYAGQVQYV
*. **:*:.*: ..::* . ..*: *:. .* :..::***:: * ::
Mvan_2594|M.vanbaalenii_PYR-1 APSQVGILPSIAFLIGVAIGGRTTLLGPVLGAIGVAWAQTLFSERFPSEW
Mflv_3800|M.gilvum_PYR-GCK APSQVGILPSIAFLIGVAIGGRATLLGPVLGAIGVAWAQTLFSERFPSAW
TH_2330|M.thermoresistible__bu APSQVGILPSIAFLIGVAIGGRTTLLGPVLGAIGVAWAQTLFSERFPSEW
MSMEG_2980|M.smegmatis_MC2_155 APSQVGIIPSIAFLIGVAIGGRTTLLGPVLGAIGVAWAQTLFSERFPSEW
MAB_2624c|M.abscessus_ATCC_199 ASPTFNIINSMLFLCAVVLGGQGNKLGVVVGAFIIVYLPNRLLGVQVQGI
*.. ..*: *: ** .*.:**: . ** *:**: :.: . : .
Mvan_2594|M.vanbaalenii_PYR-1 TYA------QGLLFIVVVGFFPAGLAGLGVLLKRRRKAGTEPPP-----E
Mflv_3800|M.gilvum_PYR-GCK TYG------QGLLFILVVGFLPAGLAGLGVFLKRRRKAEADTEP-----D
TH_2330|M.thermoresistible__bu TYA------QGLLFIVVIGFFPAGLAGLGALRSRLRRRVPDP--------
MSMEG_2980|M.smegmatis_MC2_155 TYA------QGLLFIVVVGFFPAGIAGLGVALRRWRRNKPDPTPPGPDAE
MAB_2624c|M.abscessus_ATCC_199 DLGNLKYLFFGLALVVLMLFRPQGLFPAHQKLMAYWRKTSER--------
. ** ::::: * * *: : .:
Mvan_2594|M.vanbaalenii_PYR-1 KAPDTDPDTEKVGAGS
Mflv_3800|M.gilvum_PYR-GCK KASDTDPDAEKVGASS
TH_2330|M.thermoresistible__bu AVPVKHAQEVKVEAAV
MSMEG_2980|M.smegmatis_MC2_155 PTTDSDPDAEKVGAAS
MAB_2624c|M.abscessus_ATCC_199 -LSAQKPDEVEAGA--
. ..: :. *