For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRLLACLGAVLCLVGSVSVPHAAAVPATGIVQPAGSMPIPDGPAQAWIVADMDTGAVLAARDEHGHFAPA STIKVLLAMVVLDELPLDATIVANEADTLVECNCAGVTPGRTYTTRQLLEALLLVSGNDAANTLATMLGG RDVAVMKMNAKAATLGAYGTNAGSPSGIDGPGIDIWSTPRDLAVIFRAAMADPVFAQIVAQPTAVFPAKT GDVVLVNQDELLHRYPGTFGGKTGYTDLARKTFVGGAARNGRHLVVALMYGLVKEGGPTYWDQAAALLDW GFALGPNSGVSRL*
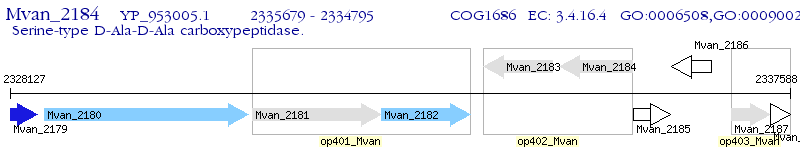
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2184 | - | - | 100% (294) | Serine-type D-Ala-D-Ala carboxypeptidase |
| M. vanbaalenii PYR-1 | Mvan_2183 | - | 2e-77 | 55.87% (281) | peptidase S11, D-alanyl-D-alanine carboxypeptidase 1 |
| M. vanbaalenii PYR-1 | Mvan_1562 | - | 1e-59 | 45.59% (272) | Serine-type D-Ala-D-Ala carboxypeptidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2935 | dacB2 | 1e-108 | 66.67% (294) | D-alanyl-D-alanine carboxypeptidase |
| M. gilvum PYR-GCK | Mflv_4179 | - | 1e-146 | 85.52% (297) | Serine-type D-Ala-D-Ala carboxypeptidase |
| M. tuberculosis H37Rv | Rv2911 | dacB2 | 1e-108 | 66.67% (294) | D-alanyl-D-alanine carboxypeptidase |
| M. leprae Br4923 | MLBr_00691 | - | 7e-59 | 45.82% (275) | putative D-alanyl-D-alanine carboxypeptidase |
| M. abscessus ATCC 19977 | MAB_3234 | - | 2e-93 | 60.29% (277) | D-alanyl-D-alanine carboxypeptidase DacB |
| M. marinum M | MMAR_1797 | dacB | 1e-109 | 66.78% (292) | D-alanyl-D-alanine carboxypeptidase DacB |
| M. avium 104 | MAV_3766 | - | 1e-104 | 65.07% (292) | D-alanyl-D-alanine carboxypeptidase |
| M. smegmatis MC2 155 | MSMEG_2433 | - | 1e-111 | 69.34% (287) | D-alanyl-D-alanine carboxypeptidase |
| M. thermoresistible (build 8) | TH_2392 | - | 1e-106 | 64.77% (298) | D-alanyl-D-alanine carboxypeptidase |
| M. ulcerans Agy99 | MUL_2045 | dacB | 1e-109 | 66.78% (292) | D-alanyl-D-alanine carboxypeptidase DacB |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2184|M.vanbaalenii_PYR-1 ----------------MTRLLACLGAVLCLVGS---VSVPHAAAVP-ATG
Mflv_4179|M.gilvum_PYR-GCK MNTTGRAGRGWIDFPVMTRLLASLGAALCLILAGSGVVSPTASAVP-VTG
MSMEG_2433|M.smegmatis_MC2_155 ----------------MWRYA--FGLVVLVVSGLITGPGSLAVPVARADA
TH_2392|M.thermoresistible__bu ----------------MGRFAGAFALLLIAVLSVVAPGAPVTDPVAWADP
Mb2935|M.bovis_AF2122/97 -----------------------MRKLMTATAALCACAVTVSAGAAWADA
Rv2911|M.tuberculosis_H37Rv -----------------------MRKLMTATAALCACAVTVSAGAAWADA
MMAR_1797|M.marinum_M -----------------------MRKVMAVAAALLT-AATISGATASADT
MUL_2045|M.ulcerans_Agy99 -----------------------MRKVMAVAAVLLT-AATISGATASADT
MAV_3766|M.avium_104 --------------MDRKLDFAAMRKLWTAAAALLL--VGACGAPTAAAD
MAB_3234|M.abscessus_ATCC_1997 -------------------MSDPMSTLRIRALAVLSVLFLSTLPVGVARA
MLBr_00691|M.leprae_Br4923 --------------MSFLRSTACLAAAVFAIVGPVALDLPTAVPVGHAEP
: .
Mvan_2184|M.vanbaalenii_PYR-1 IVQP----------------------------------------------
Mflv_4179|M.gilvum_PYR-GCK IVQP----------------------------------------------
MSMEG_2433|M.smegmatis_MC2_155 DIQQ----------------------------------------------
TH_2392|M.thermoresistible__bu EIRP----------------------------------------------
Mb2935|M.bovis_AF2122/97 DVQP----------------------------------------------
Rv2911|M.tuberculosis_H37Rv DVQP----------------------------------------------
MMAR_1797|M.marinum_M EMQP----------------------------------------------
MUL_2045|M.ulcerans_Agy99 EMQP----------------------------------------------
MAV_3766|M.avium_104 STQP----------------------------------------------
MAB_3234|M.abscessus_ATCC_1997 DMA-----------------------------------------------
MLBr_00691|M.leprae_Br4923 NAGPNAGPATCPYQVSTPPAVDSSEVPQTGNPPMPLAVPPKPVGGAALNS
Mvan_2184|M.vanbaalenii_PYR-1 AGSMPIPDGP-------AQAWIVADMDTGAVLAARDEHGHFAPASTIKVL
Mflv_4179|M.gilvum_PYR-GCK AGSVPVPDGP-------AQAWVLADMDTGAVLAARQENGRFAPASTIKIL
MSMEG_2433|M.smegmatis_MC2_155 VGSVAPPEGP-------AETWVVADMDTGQILAGRGEYVRHAPASTIKTL
TH_2392|M.thermoresistible__bu VGSVPAPTGP-------AQAWIVADLDTGEVLAGRNENTRYAPASTIKTL
Mb2935|M.bovis_AF2122/97 AGSVPIPDGP-------AQTWIVADLDSGQVLAGRDQNVAHPPASTIKVL
Rv2911|M.tuberculosis_H37Rv AGSVPIPDGP-------AQTWIVADLDSGQVLAGRDQNVAHPPASTIKVL
MMAR_1797|M.marinum_M AGSVPIPDGP-------AQTWIVADLDTGQVLAGRDQNVTHPPASTIKVL
MUL_2045|M.ulcerans_Agy99 AGSVPIPDGP-------AQTWIVADLDTGQVLAGRDQNVTHPPASTIKVL
MAV_3766|M.avium_104 AGSIPIPDGP-------AQTWIIADLDSGQVLAGRDPNVAHPPASTIKTL
MAB_3234|M.abscessus_ATCC_1997 ---APPPEGP-------AQAWLVADLDSGQILAARDPYATHAPASTIKVL
MLBr_00691|M.leprae_Br4923 CGIVTAPDTPPVPGDLSAEAWLVADLNSGAVIAARDPHGRHRPASIIKVL
. * * *::*::**:::* ::*.* . *** ** *
Mvan_2184|M.vanbaalenii_PYR-1 LAMVVLDELPLDATIVANEADTLVECNCAGVTPGRTYTTRQLLEALLLVS
Mflv_4179|M.gilvum_PYR-GCK LAMVVLDELPLDATVVANEADTRVECNCAGVAPGTTYTTRQLLEGLLLVS
MSMEG_2433|M.smegmatis_MC2_155 LALVVLDEVPLDSTIVADETDTDVECNCAGVAPGRTYTARQLLEAALLAS
TH_2392|M.thermoresistible__bu LAQVVLDEVPLDATIVATDEDTEVICNCAGVAPGHTYTTRQLLEGLLLAS
Mb2935|M.bovis_AF2122/97 LALVALDELDLNSTVVADVADTQAECNCVGVKPGRSYTARQLLDGLLLVS
Rv2911|M.tuberculosis_H37Rv LALVALDELDLNSTVVADVADTQAECNCVGVKPGRSYTARQLLDGLLLVS
MMAR_1797|M.marinum_M LALVALDELDLNSTVVADEADTHVECNCVGVKAGHTYTARQLLDGLLLVS
MUL_2045|M.ulcerans_Agy99 LALVALDELNLNSTVVADEADTHVECNCVGVKAGHTYTARQLLDGLLLVS
MAV_3766|M.avium_104 LAQVVLDEVSLDATVVADAADTQVECNCVGIKPGRSYTARQLLDALLLVS
MAB_3234|M.abscessus_ATCC_1997 LALVALDEVPMDATVVADAADAKAECNCVGIKAGQTYTARELLDAALLES
MLBr_00691|M.leprae_Br4923 LAMASINTLNLNKSVAGTAEDAAAEGTKVGVHDGGTYTINQLLHGLLMQS
** . :: : :: ::.. *: . . .*: * :** .:**.. *: *
Mvan_2184|M.vanbaalenii_PYR-1 GNDAANTLATMLGGRDVAVMKMNAKAATLGAYGTNAGSPSGIDGPGIDIW
Mflv_4179|M.gilvum_PYR-GCK GNDAANTLATMLGGRDAAVAKMNAKAAGLGAHGTRAGSPSGIDGPGIDIW
MSMEG_2433|M.smegmatis_MC2_155 GNDAANTLARMLGGPEAAVAKMNAKAAQLGARDTNVVTPSGLDAPGMPFW
TH_2392|M.thermoresistible__bu GNDAANTLARMLGGEQAAVAKMNALAGRLGAHGTTVASTSGLDVPGVPFW
Mb2935|M.bovis_AF2122/97 GNDAANTLAHMLGGQDVTVAKMNAKAATLGATSTHATTPSGLDGPGGSGA
Rv2911|M.tuberculosis_H37Rv GNDAANTLAHMLGGQDVTVAKMNAKAATLGATSTHATTPSGLDGPGGSGA
MMAR_1797|M.marinum_M GNDAANTLAQMLGGQDAAVAKMNAKAAALGAMNTHAATPSGLDGPGGSGW
MUL_2045|M.ulcerans_Agy99 GNDAANTLAQLLGGQDAAVAKMNAKAAALGAMNTHAATPSGLDGPGGSGW
MAV_3766|M.avium_104 GNDAANTLAHMLGGPEATVAKMNAKAAALGATNTHASTPSGLDGPNGSGS
MAB_3234|M.abscessus_ATCC_1997 GNDAANTLAHLLGGRQATVDKMNAKAAALGATSTHTDSPSGLDAPGMDMR
MLBr_00691|M.leprae_Br4923 GNDAAHSLAMQLGGMQQALQKINVLAAKLGGQDTRVATPSGLDGPGMS--
*****::** *** : :: *:*. *. **. .* . :.**:* *.
Mvan_2184|M.vanbaalenii_PYR-1 STPRDLAVIFRAAMADPVFAQIVAQPTAVFP--AKTGDVVLVNQDELLHR
Mflv_4179|M.gilvum_PYR-GCK STPRDLAVLFRAAMAYPVFAQITAQPTAVFP--TKSGDVVLVNQDEMLHR
MSMEG_2433|M.smegmatis_MC2_155 STPHDLAVIFRAAMADPVFAQITAMPSTVFP--AKTGDRVLVNQNELLHR
TH_2392|M.thermoresistible__bu STPHDLAVIFRAALNHPVFAEITAMPSAVFP--ARRGEMVLVNQNELLQR
Mb2935|M.bovis_AF2122/97 STAHDLVVIFRAAMANPVFAQITAEPSAMFP--SDNGEQLIVNQDELLQR
Rv2911|M.tuberculosis_H37Rv STAHDLVVIFRAAMANPVFAQITAEPSAMFP--SDNGEQLIVNQDELLQR
MMAR_1797|M.marinum_M STAHDLAVIFRTAMANPVFAHITAEPSAMFP--GEDGEHPIVNQDELLQR
MUL_2045|M.ulcerans_Agy99 STAHDLAVIFRTAMANPVFAHITAEPSAMFP--GEDGEHPIVNQDELLQR
MAV_3766|M.avium_104 STAHDLAVIFRAAMANPVFAQITAEPSAMFP--GDNGLQPITNQDELLQR
MAB_3234|M.abscessus_ATCC_1997 TSPHDLAVIFRAALANPVFAEITAQPSAPFP--GRG----LHNQNELMYR
MLBr_00691|M.leprae_Br4923 TSAYDIGLFYRYAWQNPSFSNIVATRTFNFPGPGDHPGYQLENDNQLLYQ
::. *: :::* * * *:.*.* : ** : *::::: :
Mvan_2184|M.vanbaalenii_PYR-1 YPGTFGGKTGYTDLARKTFVGGAARNGRHLVVALMYGLVKEGGPTYWDQA
Mflv_4179|M.gilvum_PYR-GCK YPGTFGGKTGYTDLARKTFVAGASRNGRHLVVAMMYGLVTEGGPTYWDQA
MSMEG_2433|M.smegmatis_MC2_155 YPGTLGGKTGFTDIARKTFVAAAQRDGRRLVIAMMYGLVKEGGPTYWDQA
TH_2392|M.thermoresistible__bu YPGALGGKTGYTDIARKTYVGAAQRDGRRLVVALMYGLVADGGPTYWDQA
Mb2935|M.bovis_AF2122/97 YPGAIGGKTGYTNAARKTFVGAAARGGRRLVIAMMYGLVKEGGPTYWDQA
Rv2911|M.tuberculosis_H37Rv YPGAIGGKTGYTNAARKTFVGAAARGGRRLVIAMMYGLVKEGGPTYWDQA
MMAR_1797|M.marinum_M YPGAIGGKTGYTNAARKTFVGAAARNGRRLVIAMMYGLVKEGGPTYWDQA
MUL_2045|M.ulcerans_Agy99 YPGAIGGKTGYTNAARKTFVGAAARNGRRLVIAMMYGLVKEGGPTYWDQA
MAV_3766|M.avium_104 YPGAIGGKTGFTNAARKTFVGAAARGGRRLVIAMMYGLIKEGGPSYWDQA
MAB_3234|M.abscessus_ATCC_1997 YPGVIGGKTGFTDIARKTYVVAAERDGKRLVVSMMYGLVHEGGPTYWDQA
MLBr_00691|M.leprae_Br4923 YPGALGGKTGYTDDAGQTFVGAANRNGRRLMAVLLHGTRQPIAP--WEQA
***.:*****:*: * :*:* .* *.*::*: :::* .* *:**
Mvan_2184|M.vanbaalenii_PYR-1 AALLDWGFALGPNSGVSRL-------------------------------
Mflv_4179|M.gilvum_PYR-GCK SALLDWGFALGPNAGVSRL-------------------------------
MSMEG_2433|M.smegmatis_MC2_155 AGLLDWGFAQDRSASIGAL-------------------------------
TH_2392|M.thermoresistible__bu TALLDWGFSQDTGTAIGVL-------------------------------
Mb2935|M.bovis_AF2122/97 ATLFDWGFALNPQASVGSL-------------------------------
Rv2911|M.tuberculosis_H37Rv ATLFDWGFALNPQASVGSL-------------------------------
MMAR_1797|M.marinum_M GSLFDWGFALNPQSGIGSL-------------------------------
MUL_2045|M.ulcerans_Agy99 GSLFDWGFALNPQSGIGSL-------------------------------
MAV_3766|M.avium_104 ATLFDWGFAQSPQASIGSL-------------------------------
MAB_3234|M.abscessus_ATCC_1997 ASLFDWGFVNDGSSSVGSL-------------------------------
MLBr_00691|M.leprae_Br4923 AHLLDYGFSTAVGTQIGTLIEPDPTVAGNADRHGHGLQAAGIMSSNDTLP
*:*:** : :. *
Mvan_2184|M.vanbaalenii_PYR-1 -----------------------------
Mflv_4179|M.gilvum_PYR-GCK -----------------------------
MSMEG_2433|M.smegmatis_MC2_155 -----------------------------
TH_2392|M.thermoresistible__bu -----------------------------
Mb2935|M.bovis_AF2122/97 -----------------------------
Rv2911|M.tuberculosis_H37Rv -----------------------------
MMAR_1797|M.marinum_M -----------------------------
MUL_2045|M.ulcerans_Agy99 -----------------------------
MAV_3766|M.avium_104 -----------------------------
MAB_3234|M.abscessus_ATCC_1997 -----------------------------
MLBr_00691|M.leprae_Br4923 VRIGVAVVGTLIVFGLIMITRSLNRRPQH