For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAHRNGFDDFAGKRCLITGAASGIGRATALKLAARGAELFLTDRDADGLALTVADARALGAVVAAHRALD ISDYDAVAGFGADIHAAHPAMDVVMNIAGVSAWGTVDKLTHHHWKSLVDINLMGPIHVIETFVPPMIEAR RGGHLVNVSSAAGIVALPWHAAYSASKYGLRGLSEVLRFDLARHRIGVSVVVPGAVRTPLVDTVHIAGVD RDDPNVRKWTDRFAGHAVSPEVAADKILRGVVKNRYLVFTSHDIRALYLFKRTAWVPYSVTMKQVNVLFT RALRPNPKQR
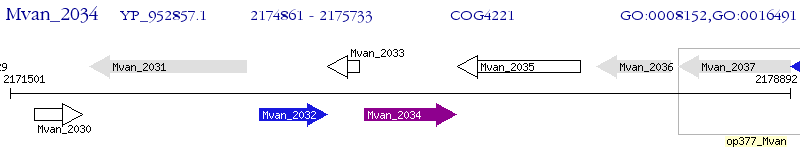
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2034 | - | - | 100% (290) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4479 | - | 8e-33 | 32.58% (264) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_3101 | - | 6e-31 | 35.94% (256) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3083c | - | 1e-127 | 76.66% (287) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4313 | - | 1e-142 | 84.83% (290) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv3057c | - | 1e-127 | 76.66% (287) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01740 | - | 1e-126 | 78.42% (278) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3420c | - | 1e-114 | 71.48% (277) | short chain dehydrogenase |
| M. marinum M | MMAR_1636 | - | 1e-129 | 79.50% (278) | short chain alcohol dehydrogenase/reductase |
| M. avium 104 | MAV_3924 | - | 1e-132 | 79.79% (287) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2291 | - | 1e-129 | 80.14% (282) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3261 | - | 3e-95 | 82.83% (198) | PUTATIVE ABC transporter ATP-binding protein |
| M. ulcerans Agy99 | MUL_3400 | - | 1e-130 | 79.86% (278) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3083c|M.bovis_AF2122/97 MLQRGAGQYFAGKRCFVTGAASGIGRATALRLAAQGAELYLTDRDRDGLA
Rv3057c|M.tuberculosis_H37Rv MLQRGAGQYFAGKRCFVTGAASGIGRATALRLAAQGAELYLTDRDRDGLA
MMAR_1636|M.marinum_M MAQR---RYFAGKRCLVTGAASGIGRATALRLAEQGAELYLTDRDGDGLA
MUL_3400|M.ulcerans_Agy99 MAQR---RYFAGKRCLVTGAASGIGRATALRLAEQGAELYLTDRDGDGLA
MAV_3924|M.avium_104 MAQKASGRYYAGKRCLVTGAASGIGRATALRLAAHGAELYLTDRDGDGLR
MLBr_01740|M.leprae_Br4923 MVKT--AQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDRHCDGLA
Mvan_2034|M.vanbaalenii_PYR-1 MAHRNGFDDFAGKRCLITGAASGIGRATALKLAARGAELFLTDRDADGLA
Mflv_4313|M.gilvum_PYR-GCK MAQRSGSDRFSGKRCFVTGAASGIGRATALKLAARGAELYLTDRDADGLA
TH_3261|M.thermoresistible__bu ----------------------------------------VVDELGDRLR
MSMEG_2291|M.smegmatis_MC2_155 MAQG---SDFAGKRCFVTGAASGIGRATALALAAAGAELYLTDRDADGLV
MAB_3420c|M.abscessus_ATCC_199 MAYE-----FTGKRCFVTGAASGIGRATALRLGREGAELFLTDRAADGLE
:.*. * *
Mb3083c|M.bovis_AF2122/97 QTVCDARALGAQVPEHRVLDVSDYQDVAAFAADIHARHPSMDVVLNIAGV
Rv3057c|M.tuberculosis_H37Rv QTVCDARALGAQVPEHRVLDVSDYQDVAAFAADIHARHPSMDVVLNIAGV
MMAR_1636|M.marinum_M QTVSAARALGAQVPEHRVLDISDYDEVAAFAADIHASHPSMDVVLNIAGI
MUL_3400|M.ulcerans_Agy99 QTVSAARALGAQVPEHRVLDISDYDEVAAFAADIHASHPSMDVVLNIAGI
MAV_3924|M.avium_104 LTVEDARALGASVPEHRALDIADYDQVASFAADIHAAHPPMDVVLNIAGV
MLBr_01740|M.leprae_Br4923 QTVSEARALGAQVPEYRALDISDYDEVAAFGADIHARHPSMDIVMNIAGV
Mvan_2034|M.vanbaalenii_PYR-1 LTVADARALGAVVAAHRALDISDYDAVAGFGADIHAAHPAMDVVMNIAGV
Mflv_4313|M.gilvum_PYR-GCK LTVADARALGGVVAEHRALDISDHDAVVAFADDIHSRHPAMDVVMNIAGI
TH_3261|M.thermoresistible__bu RX----------------------------------XHPAMDVVMNIAGV
MSMEG_2291|M.smegmatis_MC2_155 QTVADARALGAEVPAHRALDISDYDQVREFAKDIHAAHGSMDVVMNIAGV
MAB_3420c|M.abscessus_ATCC_199 SVVEEIKAAGGKVSAYRALDISDHEAVAAFASDILATHPSMDVLLNIAGI
* .**:::****:
Mb3083c|M.bovis_AF2122/97 SAWGTVDQLTHDQWSRMVAINLMGPIHVIETLVPPMVAAGRGGHLVNVSS
Rv3057c|M.tuberculosis_H37Rv SAWGTVDQLTHDQWSRMVAINLMGPIHVIETLVPPMVAAGRGGHLVNVSS
MMAR_1636|M.marinum_M SAWGTVDRLSHEHWSKMVAVNLMGPIHVIETFVPPMVAAGRGGHLVNVSS
MUL_3400|M.ulcerans_Agy99 SAWGTVDRLSHEHWSKMVAVNLMGPIHVIETFVPPMVAAGRGGHLVNVSS
MAV_3924|M.avium_104 SAWGTVDRLTHEQWRKMIDINLMGPIHVIETFVPPMVAAGRGGHLANVSS
MLBr_01740|M.leprae_Br4923 SVWGTVDRLTHDQWSNVVAINLMGPIHIIETFVPAILAAGRGGHLVNVSS
Mvan_2034|M.vanbaalenii_PYR-1 SAWGTVDKLTHHHWKSLVDINLMGPIHVIETFVPPMIEARRGGHLVNVSS
Mflv_4313|M.gilvum_PYR-GCK SAWGTVDKLTHDDWTSLININLMGPIHVIETFVPPMMEARRGGHLVNVSS
TH_3261|M.thermoresistible__bu SAWGTVDQLTHQQWKAMIDINLMGPIHVVETFIPPMMAAGRGGHLVNVSS
MSMEG_2291|M.smegmatis_MC2_155 SAWGTVDRLSHQQWRSMVDINLMGPIHVIEEFIPPMIEARRGGHLVNVSS
MAB_3420c|M.abscessus_ATCC_199 SVWGSVDRLSHQQWRSVVDINLMGPIHVIESFVPAMIKARKGGQIVNVSS
*.**:**:*:*..* :: :*******::* ::*.:: * :**::.****
Mb3083c|M.bovis_AF2122/97 AAGLVGLPWHAAYSASKYGLRGLSEVLRFDLARHGIGVSVVVPGAVKTPL
Rv3057c|M.tuberculosis_H37Rv AAGLVGLPWHAAYSASKYGLRGLSEVLRFDLARHGIGVSVVVPGAVKTPL
MMAR_1636|M.marinum_M AAGLVALPWHAAYSASKYGLRGLSEVLRFDLARHRIGVSVVVPGAVDTPL
MUL_3400|M.ulcerans_Agy99 AAGLVALPWHAAYSASKYGLRGLSEVLRFDLARHRIGVSVVVPGAVDTPL
MAV_3924|M.avium_104 AAGLVALPWHAAYSASKYGLRGLSEVLRFDLARHRIGVSVVVPGAVNTPL
MLBr_01740|M.leprae_Br4923 AAGLVALPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVVVPGAVRTPL
Mvan_2034|M.vanbaalenii_PYR-1 AAGIVALPWHAAYSASKYGLRGLSEVLRFDLARHRIGVSVVVPGAVRTPL
Mflv_4313|M.gilvum_PYR-GCK AAGIVALPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVVVPGAVRTPL
TH_3261|M.thermoresistible__bu AAGLVALPWHAAYSASKYGLRGMSEVLRFDLARHGIGVSVVVPGAVRTGL
MSMEG_2291|M.smegmatis_MC2_155 AAGLVALPWHGAYSASKFGLRGVSEVLRFDLARHRIGVSVVVPGAVKTGL
MAB_3420c|M.abscessus_ATCC_199 AAGIVALPWHAAYSASKYGLRGVSEVLRFDLARYKIGVTVVVPGAVNTPL
***:*.****.******:****:**********: ***:******* * *
Mb3083c|M.bovis_AF2122/97 VNTVEIAGVDRDDPRVNRWVERFSGHAVTPEKAADKILAGVTRNRYLVYT
Rv3057c|M.tuberculosis_H37Rv VNTVEIAGVDRDDPRVNRWVERFSGHAVTPEKAADKILAGVTRNRYLVYT
MMAR_1636|M.marinum_M VNTVEIAGVDREDPQVNRWVNRFSGHAVSPEKAAEKILAGVARNRYLVYT
MUL_3400|M.ulcerans_Agy99 VDTVEIAGVDREDPQVNRWVNRFSGHAVSPEKAAEKILAGVARNRYLVYT
MAV_3924|M.avium_104 VNTVEIAGVDRDDPDVARWVRRFAGHAVSPEKAADKILAGVARNRYLIYT
MLBr_01740|M.leprae_Br4923 VDTIEIAGVDRQDPRFSRWVDRFRGHAVSAERAAEKILAGVAKNRYLIYT
Mvan_2034|M.vanbaalenii_PYR-1 VDTVHIAGVDRDDPNVRKWTDRFAGHAVSPEVAADKILRGVVKNRYLVFT
Mflv_4313|M.gilvum_PYR-GCK VDTVRIAGVDRDDPRVQKWTNRFSGHAVSPDVAADKILDGVARNRYLIYT
TH_3261|M.thermoresistible__bu VQTVQIAGVDRDDPRVQKWVDRFSGHAVSPERAADKILAGVRRNRYLIYT
MSMEG_2291|M.smegmatis_MC2_155 VQTVEIAGVDREDPDVKKWVDRFAGHAISPEKAAAKILAGVRRNRFLIYT
MAB_3420c|M.abscessus_ATCC_199 VRTVDIAGVDRENENVARWIGRFVGHAVSPEKVADRILYGIRHNRYLVYT
* *: ******:: . :* ** ***::.: .* :** *: :**:*::*
Mb3083c|M.bovis_AF2122/97 SADIRALYAFKRYAWWPYTLVMRRVNVFFTRALRPGP-------------
Rv3057c|M.tuberculosis_H37Rv SADIRALYAFKRYAWWPYTLVMRRVNVFFTRALRPGP-------------
MMAR_1636|M.marinum_M SADIRALYAFKRLAWWPYSVAMRQVNVIFTRALKPKPVALPQRE------
MUL_3400|M.ulcerans_Agy99 SADIRALYAFKRLAWWPYSVAMRQVNVIFTRALKPKPVALPQRE------
MAV_3924|M.avium_104 SPDIRALYAFKRLAWWPYSVAMRQVNVIFTRALRPAPATLVRHDQLETHP
MLBr_01740|M.leprae_Br4923 SHDIRVLYGFKRLAWWPYGVVMKQVNVVFTRALRPSPATMRCVEPIKVGV
Mvan_2034|M.vanbaalenii_PYR-1 SHDIRALYLFKRTAWVPYSVTMKQVNVLFTRALRPNPKQR----------
Mflv_4313|M.gilvum_PYR-GCK SLDIRALYLFKRTMWLPYSIAMKQVNPLFTRALRPKTKQR----------
TH_3261|M.thermoresistible__bu SADIRALYLFKRLAWAPYSFAMRRVNVLFTRALRPGRP------------
MSMEG_2291|M.smegmatis_MC2_155 SADIRALYTFKRVAWWPYSVAMRQVNVLFTRALRSGTSPR----------
MAB_3420c|M.abscessus_ATCC_199 SVDIRALYLFKRTLWWPYSVAMRVANYAFTNALRPGFQKTGSASSPRDS-
* ***.** *** * ** ..*: .* **.**:.
Mb3083c|M.bovis_AF2122/97 --------------
Rv3057c|M.tuberculosis_H37Rv --------------
MMAR_1636|M.marinum_M --------------
MUL_3400|M.ulcerans_Agy99 --------------
MAV_3924|M.avium_104 EQPDRPVELE----
MLBr_01740|M.leprae_Br4923 YSQPRSGEGGKSGN
Mvan_2034|M.vanbaalenii_PYR-1 --------------
Mflv_4313|M.gilvum_PYR-GCK --------------
TH_3261|M.thermoresistible__bu --------------
MSMEG_2291|M.smegmatis_MC2_155 --------------
MAB_3420c|M.abscessus_ATCC_199 --------------