For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TMTALDQEVIAVHAAEEQLRAQIRDAAIAKIGAQGFRTPIRAIAAAAGLSADVVIDLYGSKRNLLKACDD HVVETIRTSKSQALQSRDPATWRAALAGIESYAPLMAYLVRSIEDGQSLADGLLDRMIENVIDYLDDGVR AGTVKPSRNPRARAKFLALSNAGGFLLYRHRHPTPRDMAAVLRDYAADMIEPALELYSSGLMADTTMLDA LAEPLARQRNGAHHRSLVPPASPDSTVNVPPTSPAR*
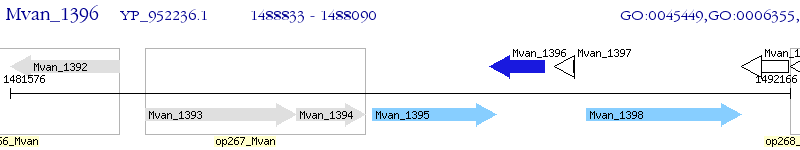
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1396 | - | - | 100% (247) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4498 | - | 5e-60 | 57.21% (201) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4277 | - | 3e-05 | 25.71% (140) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1251c | - | 4e-61 | 55.87% (213) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_2198 | - | 2e-54 | 54.00% (200) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1219c | - | 4e-61 | 55.87% (213) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1360c | - | 1e-43 | 45.83% (192) | TetR family transcriptional regulator |
| M. marinum M | MMAR_4219 | - | 1e-60 | 58.33% (204) | transcriptional regulator |
| M. avium 104 | MAV_1363 | - | 2e-59 | 53.70% (216) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_5074 | - | 6e-58 | 57.67% (189) | transcriptional regulatory protein |
| M. thermoresistible (build 8) | TH_0547 | - | 6e-62 | 56.34% (213) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_1500 | - | 1e-12 | 31.48% (162) | transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1251c|M.bovis_AF2122/97 -----------MR-SADLTAHARIREAAIEQFGRHGF-GVGLRAIAEAAG
Rv1219c|M.tuberculosis_H37Rv -----------MR-SADLTAHARIREAAIEQFGRHGF-GVGLRAIAEAAG
MMAR_4219|M.marinum_M -----------MR-SVDLTAAARIRDAAIEQFGQHGF-GVSLRAIAEGAG
MAV_1363|M.avium_104 --------MLNMR-SADLTAAARIRDAAIEQFGEHGF-GVGLRRIAEAAG
MSMEG_5074|M.smegmatis_MC2_155 --------------MDDRTAMARIRDTAIEQFGQHGF-NVGLRSIAEAAG
TH_0547|M.thermoresistible__bu -----------MRSADDRTAAARIRDAAIDQFGRHGF-DVSVRAIAEAAG
Mvan_1396|M.vanbaalenii_PYR-1 MTMTALDQEVIAVHAAEEQLRAQIRDAAIAKIGAQGF-RTPIRAIAAAAG
Mflv_2198|M.gilvum_PYR-GCK -----------MRSVDDLTAAARIRDAAVELFGAEGF-AVSVRTIAAAAG
MAB_1360c|M.abscessus_ATCC_199 -----------MRSADDLTAAARIRDAAIKLWGEQGN-GASVRAIADAAG
MUL_1500|M.ulcerans_Agy99 ----------MHLATDDRTARARIRDEALRLFAERGPDAVTMRDIATAAG
: *:**: *: . .* . :* ** .**
Mb1251c|M.bovis_AF2122/97 VSAALVIHHFGSKEGLRKACDDFVAEEIRSSKAAALKSNDPT-----TWL
Rv1219c|M.tuberculosis_H37Rv VSAALVIHHFGSKEGLRKACDDFVAEEIRSSKAAALKSNDPT-----TWL
MMAR_4219|M.marinum_M VSAALVIHHFGSKEGLRKACDNYVAEEIRSEKLTAMQSNDPA-----TWL
MAV_1363|M.avium_104 VSAALVIHHFGSKEGLRKACDDHIAEQIRESKTEALQTNDPA-----VWF
MSMEG_5074|M.smegmatis_MC2_155 VSAALVIHHFGSKEGLRKACDDHVAEVILELKTESMHASDPA-----TWL
TH_0547|M.thermoresistible__bu VSPALVIHHFGSKDNLRKACDDHIAEEIRSGKSAAIRSNDPA-----TWF
Mvan_1396|M.vanbaalenii_PYR-1 LSADVVIDLYGSKRNLLKACDDHVVETIRTSKSQALQSRDPA-----TWR
Mflv_2198|M.gilvum_PYR-GCK VSPGLVIHHFGSKDQLHKACDAYVAETVRAAKSESIQTSDPA-----TWL
MAB_1360c|M.abscessus_ATCC_199 VSPALVIHHYGSKDGLRQAVDDYLLAYIRSEKSRTITSNDPQ-----VWL
MUL_1500|M.ulcerans_Agy99 VSPALLIRHYGSKDGLIEAVDNHVIAIFEVLLTEATRETEAVGLGQAAVG
:*. ::* :*** * :* * .: . : :. .
Mb1251c|M.bovis_AF2122/97 AQMAEIESYAP----LMAYLVRSMQSGGELAKMLWQKMIDNAEEYLDEGV
Rv1219c|M.tuberculosis_H37Rv AQMAEIESYAP----LMAYLVRSMQSGGELAKMLWQKMIDNAEEYLDEGV
MMAR_4219|M.marinum_M GQLAQVESYAP----LMAYLVRSMQSGGELAMMLWQQMIDNAEEYLAVGV
MAV_1363|M.avium_104 GQLAEIEEFAP----LIAYVLRSMQTGGDLAKMLWRQMIENAEGYLEEGV
MSMEG_5074|M.smegmatis_MC2_155 AYVAEIESYAP----LMAYLVRSMQSGSELAKKFWRRMVDNAEQYLEEGV
TH_0547|M.thermoresistible__bu AQIAELESYAP----IMAYLVRSMQSGSDLAKMLWQKMIDDTRWYLDEGE
Mvan_1396|M.vanbaalenii_PYR-1 AALAGIESYAP----LMAYLVRSIEDGQSLADGLLDRMIENVIDYLDDGV
Mflv_2198|M.gilvum_PYR-GCK AQMAEIEEYAP----LMAYLVRSLQAGGELGKMLWRTMTNDVEQYLEDGV
MAB_1360c|M.abscessus_ATCC_199 DAIDEIETFAP----MVRYLLHSVQSGGEPGRAFLRRSIDDAEQYLEDGV
MUL_1500|M.ulcerans_Agy99 GLLDGLATHLPPDSAVPAYLSRMLISGGTAGPGLFARLFRLSQDTLNAMA
: : . * : *: : : * . : *
Mb1251c|M.bovis_AF2122/97 RAGTVKPSRDPRARARFLAITGGGGFLLYLQMHENPTDLRAALRDYAHDM
Rv1219c|M.tuberculosis_H37Rv RAGTVKPSRDPRARARFLAITGGGGFLLYLQMHENPTDLRAALRDYAHDM
MMAR_4219|M.marinum_M RAGTIKASRDPKARAKFLAITGAGGFLLYLQMHQTPTDLRAVLRDYARDM
MAV_1363|M.avium_104 RAGTIKPSRDPRARAKYLGITGGGGLLLYLQMHDNPTDLRAVLRDYSRDM
MSMEG_5074|M.smegmatis_MC2_155 RTGMLKPSRDPRARARFLAICGGGGFLLYLQMHDNPTDLRAVLRDYGEDM
TH_0547|M.thermoresistible__bu RAGTVKPSRDPKARARFLAISGGGSFLLYLQLHDNPTDLAAVLRDYSRDM
Mvan_1396|M.vanbaalenii_PYR-1 RAGTVKPSRNPRARAKFLALSNAGGFLLYRHRHPTPRDMAAVLRDYAADM
Mflv_2198|M.gilvum_PYR-GCK RAGTVKPSADPRARAFFLSMASGGGFLLYLQLHES-TDIRVVLRDYSEAM
MAB_1360c|M.abscessus_ATCC_199 RAGTIKPSRDPKGRALWVSLNSVGSLIIYVQMHAN-EDLREILRRYADEL
MUL_1500|M.ulcerans_Agy99 AAGTASPGADPAVRAAFLLVNDLAVLTLRPRLIEV-LGVDPLTDAGMRRW
:* ... :* ** :: : . . : : : .:
Mb1251c|M.bovis_AF2122/97 VLPSLEVYTEGLLADRAMYEAFLAEA-----QQGEAHVG-----------
Rv1219c|M.tuberculosis_H37Rv VLPSLEVYTEGLLADRAMYEAFLAEA-----QQGEAHVG-----------
MMAR_4219|M.marinum_M LLPSLEIYTEGLMADSTMYDAFLAQD-----NQGESHGH-----------
MAV_1363|M.avium_104 ILPALEIYTEGLMADRTMYDAFLAAE-----DQGDSHGN-----------
MSMEG_5074|M.smegmatis_MC2_155 MLPALEIYSHGLMADTTMYETFLHQR-----EAGVPFTGTSEEAT-----
TH_0547|M.thermoresistible__bu MLPALEIYTHGLMADSTMFDAFVARETQQLSDQGGAHDDDVAAV------
Mvan_1396|M.vanbaalenii_PYR-1 IEPALELYSSGLMADTTMLDALAEPLARQ--RNGAHHRSLVPPASPDSTV
Mflv_2198|M.gilvum_PYR-GCK VLPALELYTHGLLTDSTMYDAFVAER---AHDHSSPSTGARDA-------
MAB_1360c|M.abscessus_ATCC_199 IFPAIEVYTEGLMTDSTMLDAFAVQQ-----DKSHSKGDPK---------
MUL_1500|M.ulcerans_Agy99 AGEVFAVYRDGLVSD-----------------------------------
: :* **::*
Mb1251c|M.bovis_AF2122/97 ---------
Rv1219c|M.tuberculosis_H37Rv ---------
MMAR_4219|M.marinum_M ---------
MAV_1363|M.avium_104 ---------
MSMEG_5074|M.smegmatis_MC2_155 ---------
TH_0547|M.thermoresistible__bu ---------
Mvan_1396|M.vanbaalenii_PYR-1 NVPPTSPAR
Mflv_2198|M.gilvum_PYR-GCK ---------
MAB_1360c|M.abscessus_ATCC_199 ---------
MUL_1500|M.ulcerans_Agy99 ---------