For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDDRTAMARIRDTAIEQFGQHGFNVGLRSIAEAAGVSAALVIHHFGSKEGLRKACDDHVAEVILELKTES MHASDPATWLAYVAEIESYAPLMAYLVRSMQSGSELAKKFWRRMVDNAEQYLEEGVRTGMLKPSRDPRAR ARFLAICGGGGFLLYLQMHDNPTDLRAVLRDYGEDMMLPALEIYSHGLMADTTMYETFLHQREAGVPFTG TSEEAT
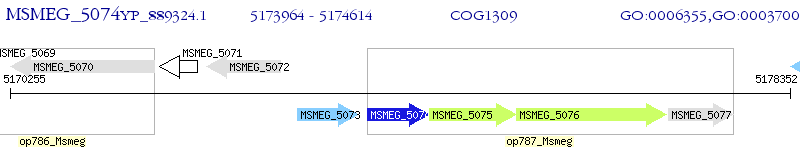
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5074 | - | - | 100% (216) | transcriptional regulatory protein |
| M. smegmatis MC2 155 | MSMEG_5282 | - | 7e-06 | 46.55% (58) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_4831 | - | 2e-05 | 28.10% (153) | transcriptional regulator, TetR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1251c | - | 1e-86 | 73.08% (208) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_2198 | - | 1e-73 | 62.33% (215) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1219c | - | 1e-86 | 73.08% (208) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1360c | - | 6e-56 | 49.50% (202) | TetR family transcriptional regulator |
| M. marinum M | MMAR_4219 | - | 5e-82 | 71.92% (203) | transcriptional regulator |
| M. avium 104 | MAV_1363 | - | 2e-85 | 72.91% (203) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_0547 | - | 2e-83 | 71.72% (198) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_1500 | - | 1e-17 | 36.54% (156) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4498 | - | 6e-87 | 70.95% (210) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2198|M.gilvum_PYR-GCK ---MRSVDDLTAAARIRDAAVELFGAEGF-AVSVRTIAAAAGVSPGLVIH
Mvan_4498|M.vanbaalenii_PYR-1 ---MRSADDLTAAARIRDAAIDLFGRDGF-AVGVRSIASAAGVSPGLVIH
Mb1251c|M.bovis_AF2122/97 ---MR-SADLTAHARIREAAIEQFGRHGF-GVGLRAIAEAAGVSAALVIH
Rv1219c|M.tuberculosis_H37Rv ---MR-SADLTAHARIREAAIEQFGRHGF-GVGLRAIAEAAGVSAALVIH
MMAR_4219|M.marinum_M ---MR-SVDLTAAARIRDAAIEQFGQHGF-GVSLRAIAEGAGVSAALVIH
MAV_1363|M.avium_104 MLNMR-SADLTAAARIRDAAIEQFGEHGF-GVGLRRIAEAAGVSAALVIH
MSMEG_5074|M.smegmatis_MC2_155 ------MDDRTAMARIRDTAIEQFGQHGF-NVGLRSIAEAAGVSAALVIH
TH_0547|M.thermoresistible__bu ---MRSADDRTAAARIRDAAIDQFGRHGF-DVSVRAIAEAAGVSPALVIH
MAB_1360c|M.abscessus_ATCC_199 ---MRSADDLTAAARIRDAAIKLWGEQGN-GASVRAIADAAGVSPALVIH
MUL_1500|M.ulcerans_Agy99 --MHLATDDRTARARIRDEALRLFAERGPDAVTMRDIATAAGVSPALLIR
* ** ****: *: :. * . :* ** .****..*:*:
Mflv_2198|M.gilvum_PYR-GCK HFGSKDQLHKACDAYVAETVRAAKSESIQTSDPA---------TWLAQMA
Mvan_4498|M.vanbaalenii_PYR-1 HFGSKDGLRKACDDFVAETVRASKSEAIQSSDPA---------TWLAQIA
Mb1251c|M.bovis_AF2122/97 HFGSKEGLRKACDDFVAEEIRSSKAAALKSNDPT---------TWLAQMA
Rv1219c|M.tuberculosis_H37Rv HFGSKEGLRKACDDFVAEEIRSSKAAALKSNDPT---------TWLAQMA
MMAR_4219|M.marinum_M HFGSKEGLRKACDNYVAEEIRSEKLTAMQSNDPA---------TWLGQLA
MAV_1363|M.avium_104 HFGSKEGLRKACDDHIAEQIRESKTEALQTNDPA---------VWFGQLA
MSMEG_5074|M.smegmatis_MC2_155 HFGSKEGLRKACDDHVAEVILELKTESMHASDPA---------TWLAYVA
TH_0547|M.thermoresistible__bu HFGSKDNLRKACDDHIAEEIRSGKSAAIRSNDPA---------TWFAQIA
MAB_1360c|M.abscessus_ATCC_199 HYGSKDGLRQAVDDYLLAYIRSEKSRTITSNDPQ---------VWLDAID
MUL_1500|M.ulcerans_Agy99 HYGSKDGLIEAVDNHVIAIFEVLLTEATRETEAVGLGQAAVGGLLDGLAT
*:***: * :* * .: . : .:.
Mflv_2198|M.gilvum_PYR-GCK EIEEYAPLMAYLVRSLQAGGELGKMLWRTMTNDVEQYLEDGVRAGTVKPS
Mvan_4498|M.vanbaalenii_PYR-1 EIEEYAPLMAYLVRTLQTGGELAKMLWQNMIDNAEQYIEEGVRAGTVKPS
Mb1251c|M.bovis_AF2122/97 EIESYAPLMAYLVRSMQSGGELAKMLWQKMIDNAEEYLDEGVRAGTVKPS
Rv1219c|M.tuberculosis_H37Rv EIESYAPLMAYLVRSMQSGGELAKMLWQKMIDNAEEYLDEGVRAGTVKPS
MMAR_4219|M.marinum_M QVESYAPLMAYLVRSMQSGGELAMMLWQQMIDNAEEYLAVGVRAGTIKAS
MAV_1363|M.avium_104 EIEEFAPLIAYVLRSMQTGGDLAKMLWRQMIENAEGYLEEGVRAGTIKPS
MSMEG_5074|M.smegmatis_MC2_155 EIESYAPLMAYLVRSMQSGSELAKKFWRRMVDNAEQYLEEGVRTGMLKPS
TH_0547|M.thermoresistible__bu ELESYAPIMAYLVRSMQSGSDLAKMLWQKMIDDTRWYLDEGERAGTVKPS
MAB_1360c|M.abscessus_ATCC_199 EIETFAPMVRYLLHSVQSGGEPGRAFLRRSIDDAEQYLEDGVRAGTIKPS
MUL_1500|M.ulcerans_Agy99 HLPPDSAVPAYLSRMLISGGTAGPGLFARLFRLSQDTLNAMAAAGTASPG
.: :.: *: : : :*. . : . : :* ...
Mflv_2198|M.gilvum_PYR-GCK ADPRARAFFLSMASGGGFLLYLQLHES-TDIRVVLRDYSEAMVLPALELY
Mvan_4498|M.vanbaalenii_PYR-1 ADPRGRARFLAMAGGGGFLLYLQMHDNPTDLRAVLRDYSEEMVLPALELY
Mb1251c|M.bovis_AF2122/97 RDPRARARFLAITGGGGFLLYLQMHENPTDLRAALRDYAHDMVLPSLEVY
Rv1219c|M.tuberculosis_H37Rv RDPRARARFLAITGGGGFLLYLQMHENPTDLRAALRDYAHDMVLPSLEVY
MMAR_4219|M.marinum_M RDPKARAKFLAITGAGGFLLYLQMHQTPTDLRAVLRDYARDMLLPSLEIY
MAV_1363|M.avium_104 RDPRARAKYLGITGGGGLLLYLQMHDNPTDLRAVLRDYSRDMILPALEIY
MSMEG_5074|M.smegmatis_MC2_155 RDPRARARFLAICGGGGFLLYLQMHDNPTDLRAVLRDYGEDMMLPALEIY
TH_0547|M.thermoresistible__bu RDPKARARFLAISGGGSFLLYLQLHDNPTDLAAVLRDYSRDMMLPALEIY
MAB_1360c|M.abscessus_ATCC_199 RDPKGRALWVSLNSVGSLIIYVQMHAN-EDLREILRRYADELIFPAIEVY
MUL_1500|M.ulcerans_Agy99 ADPAVRAAFLLVNDLAVLTLRPRLIEV-LGVDPLTDAGMRRWAGEVFAVY
** ** :: : . . : : :: .: : :*
Mflv_2198|M.gilvum_PYR-GCK THGLLTDSTMYDAFVAER-----AHDHSSPSTG-ARDA
Mvan_4498|M.vanbaalenii_PYR-1 TNGLMTDSTMYDAFVARR-----AQGIPFTSTEGAHDD
Mb1251c|M.bovis_AF2122/97 TEGLLADRAMYEAFLAEA-----QQGEAHVG-------
Rv1219c|M.tuberculosis_H37Rv TEGLLADRAMYEAFLAEA-----QQGEAHVG-------
MMAR_4219|M.marinum_M TEGLMADSTMYDAFLAQD-----NQGESHGH-------
MAV_1363|M.avium_104 TEGLMADRTMYDAFLAAE-----DQGDSHGN-------
MSMEG_5074|M.smegmatis_MC2_155 SHGLMADTTMYETFLHQR-----EAGVPFTGTSEEAT-
TH_0547|M.thermoresistible__bu THGLMADSTMFDAFVARETQQLSDQGGAHDDDVAAV--
MAB_1360c|M.abscessus_ATCC_199 TEGLMTDSTMLDAFAVQQ-----DKSHSKGDPK-----
MUL_1500|M.ulcerans_Agy99 RDGLVSD-------------------------------
.**::*