For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RVALITYSTKPRGGVVHTLNLAEALAAEGADVTVWSLARRGDTGFFREVDPSVRLRLVDFPDRPGESVTG RIVRSIDTLGARFTPHAYDIVHAQDCISANAVGTCIRTIHHLDEFTTPILAECHEKAVVAPYARICVSAA VAAEVAAGWGLHPTVIPNGVRADRFVAAAAAVDDQRRWRAELGRYVLAVGGIEPRKGTIDLLEAHHLLRQ RHPDLALVIAGGETLFDYRDYRARFERRCHELDVRPVILGAVADAELPSLVAAAEVFAFPSTKEGFGLAA MEALAAGRPVVARELPVLREVFGDTVRYASDVEGFADALAAAIGDAGRAAAGRALAESMTWEAAARAHLA FYTAHPAPAAAVP*
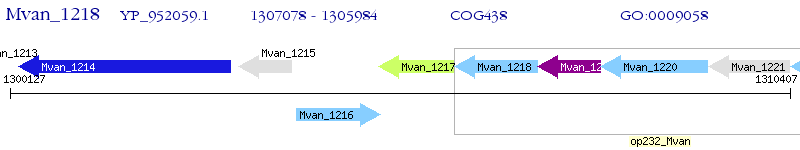
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1218 | - | - | 100% (364) | glycosyl transferase, group 1 |
| M. vanbaalenii PYR-1 | Mvan_5718 | - | 8e-15 | 26.45% (431) | glycosyl transferase, group 1 |
| M. vanbaalenii PYR-1 | Mvan_3550 | - | 8e-15 | 34.62% (234) | glycosyl transferase, group 1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2642c | pimA | 4e-16 | 33.33% (258) | alpha-mannosyltransferase PIMA |
| M. gilvum PYR-GCK | Mflv_1099 | - | 4e-15 | 27.34% (428) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv2610c | pimA | 4e-16 | 33.33% (258) | alpha-mannosyltransferase PIMA |
| M. leprae Br4923 | MLBr_00452 | - | 2e-14 | 26.55% (388) | putative glycosyltransferase |
| M. abscessus ATCC 19977 | MAB_2894c | - | 2e-16 | 27.75% (400) | phosphatidylinositol alpha-mannosyltransferase |
| M. marinum M | MMAR_2092 | pimA | 1e-16 | 29.75% (353) | alpha-mannosyltransferase PimA |
| M. avium 104 | MAV_3486 | - | 3e-18 | 28.25% (400) | phosphatidylinositol alpha-mannosyltransferase |
| M. smegmatis MC2 155 | MSMEG_0565 | - | 1e-147 | 72.80% (353) | putative glycosyl transferases group 1 |
| M. thermoresistible (build 8) | TH_1809 | - | 3e-16 | 28.69% (359) | ALPHA-MANNOSYLTRANSFERASE PIMA |
| M. ulcerans Agy99 | MUL_3252 | pimA | 1e-16 | 29.75% (353) | alpha-mannosyltransferase PimA |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2642c|M.bovis_AF2122/97 ------------------------MRIGMICPYSFDVPGGVQSHVLQLAE
Rv2610c|M.tuberculosis_H37Rv ------------------------MRIGMICPYSFDVPGGVQSHVLQLAE
MAV_3486|M.avium_104 ------------------------MRIGMVCPYSFDVPGGVQSHVLQLAE
MMAR_2092|M.marinum_M ------------------------MRIGMVCPYSFDVAGGVQSHVLQLAE
MUL_3252|M.ulcerans_Agy99 MAHVSATMVGRLVRCPPRAAAGALMRIGMVCPYSFDVAGGVQSHVLQLAE
MLBr_00452|M.leprae_Br4923 ------------------------MRIGMICPYSFDVPGGVQSHVLQLAE
TH_1809|M.thermoresistible__bu ------------------------VRIGMVCPYSFDVPGGVQAHVLQLAE
MAB_2894c|M.abscessus_ATCC_199 ------------------------MRIGMVCPYSFDVPGGVQSHVVQLAE
Mvan_1218|M.vanbaalenii_PYR-1 ------------------------MRVALIT-YSTKPRGGVV-HTLNLAE
MSMEG_0565|M.smegmatis_MC2_155 -----------------------MMRVALLT-YSTKPRGGVV-HTLYLAE
Mflv_1099|M.gilvum_PYR-GCK ------------------------MRIALLSYRSKTHCGGQGVYVRHLSR
:*:.:: * ** :. *:.
Mb2642c|M.bovis_AF2122/97 VMRTRGHLVSVLAPA-SPHAALPDYFVSGGRAVPIPYNGSVARLRFG--P
Rv2610c|M.tuberculosis_H37Rv VMRTRGHLVSVLAPA-SPHAALPDYFVSGGRAVPIPYNGSVARLRFG--P
MAV_3486|M.avium_104 VMRRRGHDVSVLAPA-SPHAVLPGYVVSAGKAVPIPYNGSVARLRFG--P
MMAR_2092|M.marinum_M VMRARGHEVSVLAPA-SPHAALPDYVVSGGKAVPIPYNGSVARLRFG--P
MUL_3252|M.ulcerans_Agy99 VMRARGHEVSVLAPA-SPHAALPDYVVSGGKAVPIPYNGSVARLRFG--P
MLBr_00452|M.leprae_Br4923 VMRARGQQVRVLAPA-SPDVSLPEYVVSAGRAIPIPYNGSVARLQFS--P
TH_1809|M.thermoresistible__bu VMRARGHEVSVLAPF-SPQVRLPDYVVSGGRAVPIPYNGSVARLRFG--P
MAB_2894c|M.abscessus_ATCC_199 VMRDRGHHVSLLAPS-SSDLELPEFVVSGGKAVPIPYNGSVARLRFG--P
Mvan_1218|M.vanbaalenii_PYR-1 ALAAEGADVTVWSLARRGDTGFFREVDPSVRLRLVDFPDRPGESVTGRIV
MSMEG_0565|M.smegmatis_MC2_155 AMARLGVDVTVWSLARSGDRGFFREINPAVTVRLVEFVDHPGDTVTDRIL
Mflv_1099|M.gilvum_PYR-GCK GLVELGHDVEVFSGQPYPEILDPRVRLTEVPSLDLYREPDPFRIPRPSEI
: * * : : . . :
Mb2642c|M.bovis_AF2122/97 ATHRKVKKWLA-----------------------HGDFDVLHLHEPNAPS
Rv2610c|M.tuberculosis_H37Rv ATHRKVKKWLA-----------------------HGDFDVLHLHEPNAPS
MAV_3486|M.avium_104 ATHRKVKKWLV-----------------------DGDFDVLHLHEPNAPS
MMAR_2092|M.marinum_M ATHRKVKKWLS-----------------------EGDFDVLHLHEPNAPS
MUL_3252|M.ulcerans_Agy99 ATHRKVKKWLS-----------------------EGDFDVLHLHEPNAPS
MLBr_00452|M.leprae_Br4923 AVHSRVRRWLV-----------------------DGDFDVLHLHEPNAPS
TH_1809|M.thermoresistible__bu ATHRKVKKWLA-----------------------EGQFDVLHLHEPNAPS
MAB_2894c|M.abscessus_ATCC_199 ATYAKTRRWLV-----------------------DGDFDVLHLHEPNAPS
Mvan_1218|M.vanbaalenii_PYR-1 RSIDTLGARFT-----------------------PHAYDIVHAQDCISAN
MSMEG_0565|M.smegmatis_MC2_155 RSIETLRDAFT-----------------------PADYDIVHAQDCISAN
Mflv_1099|M.gilvum_PYR-GCK RSSIDLEELLTTWTAGFPEPKTFSMRAARLLAARRGDFDVVHDNQCLGTG
: :*::* :: ...
Mb2642c|M.bovis_AF2122/97 LSMLALNIAEGPIVATFHTSTTKSLTLTVFQGILR------------PMH
Rv2610c|M.tuberculosis_H37Rv LSMLALNIAEGPIVATFHTSTTKSLTLTVFQGILR------------PMH
MAV_3486|M.avium_104 LSMLALNIAEGPIVATFHTSTTKSLTLTVFQGILR------------PMH
MMAR_2092|M.marinum_M LSMLALNIAEGPIVATFHTSTTKSLTLTVFQSILR------------PMH
MUL_3252|M.ulcerans_Agy99 LSMLALNIAEGPIVATFHTSTTKSLTLTVFQSILR------------PMH
MLBr_00452|M.leprae_Br4923 LSMWALRVAEGPIVATFHTSTTKSLTLSVFQGVLR------------PWH
TH_1809|M.thermoresistible__bu LSMLALQAAEGPIVATFHTSTTKSLTLSVFQGILR------------PYH
MAB_2894c|M.abscessus_ATCC_199 LSMLALMVAEGPIVATFHTSTTKSLTLSVFQAVLR------------PWH
Mvan_1218|M.vanbaalenii_PYR-1 --------AVGTCIRTIHH--LDEFTTPILAECHE------------KAV
MSMEG_0565|M.smegmatis_MC2_155 --------AVGPCIRTIHH--LDQFKTPVLAECHE------------KAI
Mflv_1099|M.gilvum_PYR-GCK --LLTIAESGMPVVATVHHPITRDKVLEVAAAKWWRKPLVRRWYGFAEMQ
: . : *.* . :
Mb2642c|M.bovis_AF2122/97 EKIVGRIAVSDLARRWQMEALGSDAVEIPNGVDVDSFASAARLDGYPR--
Rv2610c|M.tuberculosis_H37Rv EKIVGRIAVSDLARRWQMEALGSDAVEIPNGVDVDSFASAARLDGYPR--
MAV_3486|M.avium_104 EKIVGRIAVSDLARRWQMEALGTDAVEIPNGVDVASFASAPRLDGYPR--
MMAR_2092|M.marinum_M EKIVGRIAVSDLARRWQMEALGSDAVEIPNGVDVGSFASSSCLDGYPR--
MUL_3252|M.ulcerans_Agy99 EKIVGRIAVSDLARRWQMEALGSDAVEIPNGVDVGSFASSSCLDGYPR--
MLBr_00452|M.leprae_Br4923 EKIIGRIAVSDLARRWQMEALGSDAVEIPNGVNVDSLSSAPQLAGYPR--
TH_1809|M.thermoresistible__bu EKIVGRIAVSDLARRWQMEALGSDAVEIPNGVDVASFAEAPLLDGYPR--
MAB_2894c|M.abscessus_ATCC_199 EKIVGRIAVSELARRWQMEALGSDAVEIPNGVDVRSFSGAPVLEGYPR--
Mvan_1218|M.vanbaalenii_PYR-1 VAPYARICVSAAVAAEVAAGWGLHPTVIPNGVRADRFVAAAAAVDDQRRW
MSMEG_0565|M.smegmatis_MC2_155 AEPYARICVSAAVAAEVRAGWGFDPVVIPNGVDAQRFSAAAAQEAGIRRW
Mflv_1099|M.gilvum_PYR-GCK KDVARKIPELVTVSSTSAADIAEDFGVTPEQLNVVPLGVDTQLFQPAEHR
:* . . . *: : . : . .
Mb2642c|M.bovis_AF2122/97 ---QGKTVLFLGRYDEPRKGMAVLLDALPKVVQRFPDVQLLIVG------
Rv2610c|M.tuberculosis_H37Rv ---QGKTVLFLGRYDEPRKGMAVLLDALPKVVQRFPDVQLLIVG------
MAV_3486|M.avium_104 ---AGKTVLFLGRYDEPRKGMAVLLDALPRLVERFADVQLLVVG------
MMAR_2092|M.marinum_M ---PGKTVLFLGRYDEPRKGMSVLLEALPKLVERFSDVQLLIVG------
MUL_3252|M.ulcerans_Agy99 ---PGKTVLFLGRYDEPRKGMSVLLEALPKLVERFIDVQLLIVG------
MLBr_00452|M.leprae_Br4923 ---LGKTVLFLGRYDEPRKGMSVLLDALPGVMECFDDVQLLIVG------
TH_1809|M.thermoresistible__bu ---PGRTVLFLGRYDEPRKGMAVLLGALPALVGRYPEIEILIVG------
MAB_2894c|M.abscessus_ATCC_199 ---EGKTVLFLGRYDEPRKGMDVLMGALPKIATSFPDVEILIVG------
Mvan_1218|M.vanbaalenii_PYR-1 RAELGRYVLAVGGIE-PRKGTIDLLEAHHLLRQRHPDLALVIAGGETLFD
MSMEG_0565|M.smegmatis_MC2_155 RDELGSYVLTVGGIE-PRKGTLDLVEAYAMLRQRHPEVSLVIGGGETLFD
Mflv_1099|M.gilvum_PYR-GCK ---VRNRIIAIASADVPLKGVSHLLHAVARLRVERDLELQLVAKLEP---
:: :. : * ** *: * : ::
Mb2642c|M.bovis_AF2122/97 -HGDADQLRGQAGRLAAHLRFLGQVDDAGKASAMRSADVYCAPNTGGESF
Rv2610c|M.tuberculosis_H37Rv -HGDADQLRGQAGRLAAHLRFLGQVDDAGKASAMRSADVYCAPNTGGESF
MAV_3486|M.avium_104 -RGDEDQLRTQAGELVKHIRFLGQVDDAGKASAMRSADVYCAPNLGGESF
MMAR_2092|M.marinum_M -RGDEDELRDEAGELAEHMRFLGQVDDAGKASAMRSADVYCAPNTGGESF
MUL_3252|M.ulcerans_Agy99 -RGDEDELRDEAGELAEHMRFLGQVDDAGKASAMRSADVYCAPNTGGESF
MLBr_00452|M.leprae_Br4923 -RGDEEQLRSQAGGLVEHIRFLGQVDDAGKAAAMRSADVYCAPNIGGESF
TH_1809|M.thermoresistible__bu -RGDEQELRGKAGRYARHLRFLGQVDDATKASAMRSADVYCAPHIGGESF
MAB_2894c|M.abscessus_ATCC_199 -RGDEAELRTQAGPLAKHLKFLGQVDDATKASAMRSADVYCAPNIGGESF
Mvan_1218|M.vanbaalenii_PYR-1 YRDYRARFERRCHELDVRPVILGAVADAELPSLVAAAEVFAFPST-KEGF
MSMEG_0565|M.smegmatis_MC2_155 YRDYRSAFERRCTELGVEPVILGAVADDELPGLVAGCDVFAFPST-KEGF
Mflv_1099|M.gilvum_PYR-GCK -NGPTEKLIAELG-ISDIVHISSGLSDSELAGLLASAEVACIPSL-YEGF
.. : . : . : * .. : ..:* . * *.*
Mb2642c|M.bovis_AF2122/97 GIVLVEAMAAGTAVVASDLDAFRRVLRDGEVGHLVPVDPPDLQAAALADG
Rv2610c|M.tuberculosis_H37Rv GIVLVEAMAAGTAVVASDLDAFRRVLRDGEVGHLVPVDPPDLQAAALADG
MAV_3486|M.avium_104 GIVLVEAMAAGTPVVASDLDAFRRVLRDGEVGALVPVG----DGDALADA
MMAR_2092|M.marinum_M GIVLVEAMAAGTPVVASDLDAFRRVLRDGEIGRLVPVD----DSDALAAA
MUL_3252|M.ulcerans_Agy99 GIVLVEAMAAGTPVVASDLDAFRRVLRDGEIGRLVPVD----DSDALAAA
MLBr_00452|M.leprae_Br4923 GIVLVEAMAAGTPVVASDLDAFRRVLRDGEVGHLVPAG----DSAALADA
TH_1809|M.thermoresistible__bu GIVLVEAMAAGTAVVASELDAFRRVLLDGQAGRLVPVD----DPAALAQG
MAB_2894c|M.abscessus_ATCC_199 GIVLVEAMAAGTAVVASSLDAFRRVLLDGTAGRLTPTG----DPDALAAA
Mvan_1218|M.vanbaalenii_PYR-1 GLAAMEALAAGRPVVARELPVLREVFGDTVRYASDVEG--------FADA
MSMEG_0565|M.smegmatis_MC2_155 GLAAMEALAAGRPVVTRDLPVLREVFGDTVGFAADPAG--------FARE
Mflv_1099|M.gilvum_PYR-GCK SLPAVEAMASGTPIVASRAGALPEVVGADGQCARLVKP---ADVAELTSV
.: :**:*:* .:*: .: .*. ::
Mb2642c|M.bovis_AF2122/97 LIAVLENDVLRERYVAAG-NAAVRRYDWSVVASQIMRVYETVAGSGAKVQ
Rv2610c|M.tuberculosis_H37Rv LIAVLENDVLRERYVAAG-NAAVRRYDWSVVASQIMRVYETVAGSGAKVQ
MAV_3486|M.avium_104 LIAVLEDDVLRDGYVAAG-QAAVQRYDWSVVASQIMRVYETVAATGAKVE
MMAR_2092|M.marinum_M LIEVLDSQELRERYVAAG-TEAVRRYDWSVVASQIMRVYETVAGAGVKVQ
MUL_3252|M.ulcerans_Agy99 LIEVLDSQELRERYVAAG-TEAVRRYDWSVVASQIMRVYETVAGAGVKVQ
MLBr_00452|M.leprae_Br4923 LVALLRNDVLRERYVAAG-AEAVRRYDWSVVASQIMRVYETVATSGSKVQ
TH_1809|M.thermoresistible__bu LLEVLGDDALRQRYIEVA-AEAVRRYDWSVVAGQIMRVYETVASAGAKVR
MAB_2894c|M.abscessus_ATCC_199 LIDVLGNDEGRAQLVAAG-TEAVQRYDWSVVAQQIMLVYETVTASGARVK
Mvan_1218|M.vanbaalenii_PYR-1 LAAAIGDAG----RAAAG-RALAESMTWEAAARAHLAFYTAHPAPAAAVP
MSMEG_0565|M.smegmatis_MC2_155 MTAALEEPA----DPAPG-RTLAAAMTWTQAAKTHVNFYRTLRTQDGPHP
Mflv_1099|M.gilvum_PYR-GCK LGELLDSPRELRRLGDNGRRRALEVFSWESVAAQTVAVYERACERVATC-
: : . . * .* : .*
Mb2642c|M.bovis_AF2122/97 VAS-
Rv2610c|M.tuberculosis_H37Rv VAS-
MAV_3486|M.avium_104 VAS-
MMAR_2092|M.marinum_M VAS-
MUL_3252|M.ulcerans_Agy99 VAS-
MLBr_00452|M.leprae_Br4923 VAS-
TH_1809|M.thermoresistible__bu VAT-
MAB_2894c|M.abscessus_ATCC_199 VDSW
Mvan_1218|M.vanbaalenii_PYR-1 ----
MSMEG_0565|M.smegmatis_MC2_155 ----
Mflv_1099|M.gilvum_PYR-GCK ----