For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRVALLTYSTKPRGGVVHTLYLAEAMARLGVDVTVWSLARSGDRGFFREINPAVTVRLVEFVDHPGDTVT DRILRSIETLRDAFTPADYDIVHAQDCISANAVGPCIRTIHHLDQFKTPVLAECHEKAIAEPYARICVSA AVAAEVRAGWGFDPVVIPNGVDAQRFSAAAAQEAGIRRWRDELGSYVLTVGGIEPRKGTLDLVEAYAMLR QRHPEVSLVIGGGETLFDYRDYRSAFERRCTELGVEPVILGAVADDELPGLVAGCDVFAFPSTKEGFGLA AMEALAAGRPVVTRDLPVLREVFGDTVGFAADPAGFAREMTAALEEPADPAPGRTLAAAMTWTQAAKTHV NFYRTLRTQDGPHP*
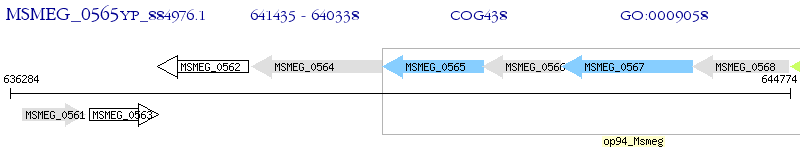
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0565 | - | - | 100% (365) | putative glycosyl transferases group 1 |
| M. smegmatis MC2 155 | MSMEG_6484 | - | 7e-19 | 26.03% (438) | glycosyl transferase |
| M. smegmatis MC2 155 | MSMEG_2935 | - | 2e-16 | 29.58% (355) | phosphatidylinositol alpha-mannosyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2642c | pimA | 2e-16 | 29.07% (399) | alpha-mannosyltransferase PIMA |
| M. gilvum PYR-GCK | Mflv_1099 | - | 5e-17 | 26.85% (391) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv2610c | pimA | 2e-16 | 29.07% (399) | alpha-mannosyltransferase PIMA |
| M. leprae Br4923 | MLBr_00452 | - | 3e-17 | 30.00% (400) | putative glycosyltransferase |
| M. abscessus ATCC 19977 | MAB_2894c | - | 8e-17 | 28.61% (395) | phosphatidylinositol alpha-mannosyltransferase |
| M. marinum M | MMAR_2092 | pimA | 5e-19 | 29.11% (395) | alpha-mannosyltransferase PimA |
| M. avium 104 | MAV_3486 | - | 2e-19 | 29.02% (386) | phosphatidylinositol alpha-mannosyltransferase |
| M. thermoresistible (build 8) | TH_0088 | - | 1e-16 | 25.24% (424) | glycosyl transferase |
| M. ulcerans Agy99 | MUL_3252 | pimA | 4e-19 | 29.04% (396) | alpha-mannosyltransferase PimA |
| M. vanbaalenii PYR-1 | Mvan_1218 | - | 1e-147 | 72.80% (353) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2642c|M.bovis_AF2122/97 ------------------------MRIGMICPYSFDVPGGVQSHVLQLAE
Rv2610c|M.tuberculosis_H37Rv ------------------------MRIGMICPYSFDVPGGVQSHVLQLAE
MMAR_2092|M.marinum_M ------------------------MRIGMVCPYSFDVAGGVQSHVLQLAE
MUL_3252|M.ulcerans_Agy99 MAHVSATMVGRLVRCPPRAAAGALMRIGMVCPYSFDVAGGVQSHVLQLAE
MAV_3486|M.avium_104 ------------------------MRIGMVCPYSFDVPGGVQSHVLQLAE
MLBr_00452|M.leprae_Br4923 ------------------------MRIGMICPYSFDVPGGVQSHVLQLAE
MAB_2894c|M.abscessus_ATCC_199 ------------------------MRIGMVCPYSFDVPGGVQSHVVQLAE
Mflv_1099|M.gilvum_PYR-GCK ------------------------MRIALLSYRSKTHCGGQGVYVRHLSR
TH_0088|M.thermoresistible__bu ------------------------MRIALLSYRSKTHCGGQGVYVRHLSR
MSMEG_0565|M.smegmatis_MC2_155 -----------------------MMRVALLTYSTKPRGG--VVHTLYLAE
Mvan_1218|M.vanbaalenii_PYR-1 ------------------------MRVALITYSTKPRGG--VVHTLNLAE
**:.:: : * :. *:.
Mb2642c|M.bovis_AF2122/97 VMRTRGHLVSVLAPA------------SPHAALPDYFVSGGRAVPIP---
Rv2610c|M.tuberculosis_H37Rv VMRTRGHLVSVLAPA------------SPHAALPDYFVSGGRAVPIP---
MMAR_2092|M.marinum_M VMRARGHEVSVLAPA------------SPHAALPDYVVSGGKAVPIP---
MUL_3252|M.ulcerans_Agy99 VMRARGHEVSVLAPA------------SPHAALPDYVVSGGKAVPIP---
MAV_3486|M.avium_104 VMRRRGHDVSVLAPA------------SPHAVLPGYVVSAGKAVPIP---
MLBr_00452|M.leprae_Br4923 VMRARGQQVRVLAPA------------SPDVSLPEYVVSAGRAIPIP---
MAB_2894c|M.abscessus_ATCC_199 VMRDRGHHVSLLAPS------------SSDLELPEFVVSGGKAVPIP---
Mflv_1099|M.gilvum_PYR-GCK GLVELGHDVEVFSGQPYPEILDPRVRLTEVPSLDLYREPDPFRIPRPSEI
TH_0088|M.thermoresistible__bu GLVELGHQVEVFSGQPYPEGLDPRVKLTKVPSLDLYREPDPFRIPRPSEI
MSMEG_0565|M.smegmatis_MC2_155 AMARLGVDVTVWSLAR--------------SGDRGFFREINPAVTVR---
Mvan_1218|M.vanbaalenii_PYR-1 ALAAEGADVTVWSLAR--------------RGDTGFFREVDPSVRLR---
: * * : : : :
Mb2642c|M.bovis_AF2122/97 ---------YNGSVARLRFGPATHRKVKKWLAHGDFDVLHLHEPNAPSLS
Rv2610c|M.tuberculosis_H37Rv ---------YNGSVARLRFGPATHRKVKKWLAHGDFDVLHLHEPNAPSLS
MMAR_2092|M.marinum_M ---------YNGSVARLRFGPATHRKVKKWLSEGDFDVLHLHEPNAPSLS
MUL_3252|M.ulcerans_Agy99 ---------YNGSVARLRFGPATHRKVKKWLSEGDFDVLHLHEPNAPSLS
MAV_3486|M.avium_104 ---------YNGSVARLRFGPATHRKVKKWLVDGDFDVLHLHEPNAPSLS
MLBr_00452|M.leprae_Br4923 ---------YNGSVARLQFSPAVHSRVRRWLVDGDFDVLHLHEPNAPSLS
MAB_2894c|M.abscessus_ATCC_199 ---------YNGSVARLRFGPATYAKTRRWLVDGDFDVLHLHEPNAPSLS
Mflv_1099|M.gilvum_PYR-GCK RSSIDLEELLTTWTAGFPEPKTFSMRAARLLAARRGDFDVVHDNQCLGTG
TH_0088|M.thermoresistible__bu RDRIDLLELLTTWTAGFPEPKTFTMRMARILAERRDEFDVVHDNQSLGTG
MSMEG_0565|M.smegmatis_MC2_155 ---------LVEFVD--HPGDTVTDRILRSIETLRDAFTPADYDIVHAQD
Mvan_1218|M.vanbaalenii_PYR-1 ---------LVDFPD--RPGESVTGRIVRSIDTLGARFTPHAYDIVHAQD
: : : : . . .
Mb2642c|M.bovis_AF2122/97 MLALNIAEGPIVATFHTSTTKSLTLTVFQGILR------------PMHEK
Rv2610c|M.tuberculosis_H37Rv MLALNIAEGPIVATFHTSTTKSLTLTVFQGILR------------PMHEK
MMAR_2092|M.marinum_M MLALNIAEGPIVATFHTSTTKSLTLTVFQSILR------------PMHEK
MUL_3252|M.ulcerans_Agy99 MLALNIAEGPIVATFHTSTTKSLTLTVFQSILR------------PMHEK
MAV_3486|M.avium_104 MLALNIAEGPIVATFHTSTTKSLTLTVFQGILR------------PMHEK
MLBr_00452|M.leprae_Br4923 MWALRVAEGPIVATFHTSTTKSLTLSVFQGVLR------------PWHEK
MAB_2894c|M.abscessus_ATCC_199 MLALMVAEGPIVATFHTSTTKSLTLSVFQAVLR------------PWHEK
Mflv_1099|M.gilvum_PYR-GCK LLTIAESGMPVVATVHHPITRDKVLEVAAAKWWRKPLVRRWYGFAEMQKD
TH_0088|M.thermoresistible__bu LLELADLGLPVVATVHHPITRDKVVEVAAAKWWRKPLVRRWYGFAEMQKR
MSMEG_0565|M.smegmatis_MC2_155 CISANAVGPCIRTIHHLDQFKTPVLAECHEKAI---------------AE
Mvan_1218|M.vanbaalenii_PYR-1 CISANAVGTCIRTIHHLDEFTTPILAECHEKAV---------------VA
: : * :
Mb2642c|M.bovis_AF2122/97 IVGRIAVSDLARRWQMEALGSDAVEIPNGVDVDSFASAARLDG-----YP
Rv2610c|M.tuberculosis_H37Rv IVGRIAVSDLARRWQMEALGSDAVEIPNGVDVDSFASAARLDG-----YP
MMAR_2092|M.marinum_M IVGRIAVSDLARRWQMEALGSDAVEIPNGVDVGSFASSSCLDG-----YP
MUL_3252|M.ulcerans_Agy99 IVGRIAVSDLARRWQMEALGSDAVEIPNGVDVGSFASSSCLDG-----YP
MAV_3486|M.avium_104 IVGRIAVSDLARRWQMEALGTDAVEIPNGVDVASFASAPRLDG-----YP
MLBr_00452|M.leprae_Br4923 IIGRIAVSDLARRWQMEALGSDAVEIPNGVNVDSLSSAPQLAG-----YP
MAB_2894c|M.abscessus_ATCC_199 IVGRIAVSELARRWQMEALGSDAVEIPNGVDVRSFSGAPVLEG-----YP
Mflv_1099|M.gilvum_PYR-GCK VARKIPELVTVSSTSAADIAEDFGVTPEQLNVVPLGVDTQLFQPA---EH
TH_0088|M.thermoresistible__bu VARRIPELITVSSSSAADISADFGVSPGQLHVVPLGVDTELFTPS---EH
MSMEG_0565|M.smegmatis_MC2_155 PYARICVSAAVAAEVRAGWGFDPVVIPNGVDAQRFSAAAAQEAGIRRWRD
Mvan_1218|M.vanbaalenii_PYR-1 PYARICVSAAVAAEVAAGWGLHPTVIPNGVRADRFVAAAAAVDDQRRWRA
:* . . . * : . : .
Mb2642c|M.bovis_AF2122/97 RQGKTVLFLGRYDEPRKGMAVLLDALPKV-VQRFPDVQLLIVG-------
Rv2610c|M.tuberculosis_H37Rv RQGKTVLFLGRYDEPRKGMAVLLDALPKV-VQRFPDVQLLIVG-------
MMAR_2092|M.marinum_M RPGKTVLFLGRYDEPRKGMSVLLEALPKL-VERFSDVQLLIVG-------
MUL_3252|M.ulcerans_Agy99 RPGKTVLFLGRYDEPRKGMSVLLEALPKL-VERFIDVQLLIVG-------
MAV_3486|M.avium_104 RAGKTVLFLGRYDEPRKGMAVLLDALPRL-VERFADVQLLVVG-------
MLBr_00452|M.leprae_Br4923 RLGKTVLFLGRYDEPRKGMSVLLDALPGV-MECFDDVQLLIVG-------
MAB_2894c|M.abscessus_ATCC_199 REGKTVLFLGRYDEPRKGMDVLMGALPKI-ATSFPDVEILIVG-------
Mflv_1099|M.gilvum_PYR-GCK RVRNRIIAIASADVPLKGVSHLLHAVARLRVERDLELQLVAKLE-----P
TH_0088|M.thermoresistible__bu RVPGRIIAIASADVPLKGISHLLHAVARLRAKRNLDVQLVSKLE-----P
MSMEG_0565|M.smegmatis_MC2_155 ELGSYVLTVGGIE-PRKGTLDLVEAYAML-RQRHPEVSLVIGGGETLFDY
Mvan_1218|M.vanbaalenii_PYR-1 ELGRYVLAVGGIE-PRKGTIDLLEAHHLL-RQRHPDLALVIAGGETLFDY
. :: :. : * ** *: * : :: ::
Mb2642c|M.bovis_AF2122/97 HGDADQLRGQAGRLAAHLRFLGQVDDAGKASAMRSADVYCAPNTGGESFG
Rv2610c|M.tuberculosis_H37Rv HGDADQLRGQAGRLAAHLRFLGQVDDAGKASAMRSADVYCAPNTGGESFG
MMAR_2092|M.marinum_M RGDEDELRDEAGELAEHMRFLGQVDDAGKASAMRSADVYCAPNTGGESFG
MUL_3252|M.ulcerans_Agy99 RGDEDELRDEAGELAEHMRFLGQVDDAGKASAMRSADVYCAPNTGGESFG
MAV_3486|M.avium_104 RGDEDQLRTQAGELVKHIRFLGQVDDAGKASAMRSADVYCAPNLGGESFG
MLBr_00452|M.leprae_Br4923 RGDEEQLRSQAGGLVEHIRFLGQVDDAGKAAAMRSADVYCAPNIGGESFG
MAB_2894c|M.abscessus_ATCC_199 RGDEAELRTQAGPLAKHLKFLGQVDDATKASAMRSADVYCAPNIGGESFG
Mflv_1099|M.gilvum_PYR-GCK NGPTEKLIAELG-ISDIVHISSGLSDSELAGLLASAEVACIPSLY-EGFS
TH_0088|M.thermoresistible__bu NGPTEKLIAELG-ISDIVQASSGLSDSELARLLASAEVACIPSLY-EGFS
MSMEG_0565|M.smegmatis_MC2_155 RDYRSAFERRCTELGVEPVILGAVADDELPGLVAGCDVFAFPSTK-EGFG
Mvan_1218|M.vanbaalenii_PYR-1 RDYRARFERRCHELDVRPVILGAVADAELPSLVAAAEVFAFPSTK-EGFG
.. : . : . : * . : ..:* . *. *.*.
Mb2642c|M.bovis_AF2122/97 IVLVEAMAAGTAVVASDLDAFRRVLR-DGEVGHLVPVDPPDLQAAALADG
Rv2610c|M.tuberculosis_H37Rv IVLVEAMAAGTAVVASDLDAFRRVLR-DGEVGHLVPVDPPDLQAAALADG
MMAR_2092|M.marinum_M IVLVEAMAAGTPVVASDLDAFRRVLR-DGEIGRLVPVD----DSDALAAA
MUL_3252|M.ulcerans_Agy99 IVLVEAMAAGTPVVASDLDAFRRVLR-DGEIGRLVPVD----DSDALAAA
MAV_3486|M.avium_104 IVLVEAMAAGTPVVASDLDAFRRVLR-DGEVGALVPVG----DGDALADA
MLBr_00452|M.leprae_Br4923 IVLVEAMAAGTPVVASDLDAFRRVLR-DGEVGHLVPAG----DSAALADA
MAB_2894c|M.abscessus_ATCC_199 IVLVEAMAAGTAVVASSLDAFRRVLL-DGTAGRLTPTG----DPDALAAA
Mflv_1099|M.gilvum_PYR-GCK LPAVEAMASGTPIVASRAGALPEVVGADGQCARLVKPA----DVAELTSV
TH_0088|M.thermoresistible__bu LPAVEAMASGTPIVASRAGALPEVLGPDGECARLVPPA----DVDALTRV
MSMEG_0565|M.smegmatis_MC2_155 LAAMEALAAGRPVVTRDLPVLREVFG-----DTVGFAA----DPAGFARE
Mvan_1218|M.vanbaalenii_PYR-1 LAAMEALAAGRPVVARELPVLREVFG-----DTVRYAS----DVEGFADA
: :**:*:* .:*: .: .*. : : ::
Mb2642c|M.bovis_AF2122/97 LIAVLENDVLRERYVAAG-NAAVRRYDWSVVASQIMRVYETVAGSGAKVQ
Rv2610c|M.tuberculosis_H37Rv LIAVLENDVLRERYVAAG-NAAVRRYDWSVVASQIMRVYETVAGSGAKVQ
MMAR_2092|M.marinum_M LIEVLDSQELRERYVAAG-TEAVRRYDWSVVASQIMRVYETVAGAGVKVQ
MUL_3252|M.ulcerans_Agy99 LIEVLDSQELRERYVAAG-TEAVRRYDWSVVASQIMRVYETVAGAGVKVQ
MAV_3486|M.avium_104 LIAVLEDDVLRDGYVAAG-QAAVQRYDWSVVASQIMRVYETVAATGAKVE
MLBr_00452|M.leprae_Br4923 LVALLRNDVLRERYVAAG-AEAVRRYDWSVVASQIMRVYETVATSGSKVQ
MAB_2894c|M.abscessus_ATCC_199 LIDVLGNDEGRAQLVAAG-TEAVQRYDWSVVAQQIMLVYETVTASGARVK
Mflv_1099|M.gilvum_PYR-GCK LGELLDSPRELRRLGDNGRRRALEVFSWESVAAQTVAVYERACERVATC-
TH_0088|M.thermoresistible__bu LGELLDSPHQRSRLGAAGRQRALDVFSWESVAAQTVSIYER-----AIC-
MSMEG_0565|M.smegmatis_MC2_155 MTAALEEPAD----PAPG-RTLAAAMTWTQAAKTHVNFYRTLRTQDGPHP
Mvan_1218|M.vanbaalenii_PYR-1 LAAAIGDAGR----AAAG-RALAESMTWEAAARAHLAFYTAHPAPAAAVP
: : . * * .* : .*
Mb2642c|M.bovis_AF2122/97 VAS-
Rv2610c|M.tuberculosis_H37Rv VAS-
MMAR_2092|M.marinum_M VAS-
MUL_3252|M.ulcerans_Agy99 VAS-
MAV_3486|M.avium_104 VAS-
MLBr_00452|M.leprae_Br4923 VAS-
MAB_2894c|M.abscessus_ATCC_199 VDSW
Mflv_1099|M.gilvum_PYR-GCK ----
TH_0088|M.thermoresistible__bu ----
MSMEG_0565|M.smegmatis_MC2_155 ----
Mvan_1218|M.vanbaalenii_PYR-1 ----