For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NYPRDMVGYGRTPPDPQWPGGAAIAVQFVLNYEEGAENCVLDGDPASETFLSEITPAEAFPNRHMSMESL YEYGSRAGLWRVLRVFERHDIPLTVFAVARALQRNPEAVAAFTELGHEIACHGLRWKSYQLTDRDTERVH LAEAVQILQDLTGSTPSGWYTGRDSPHTRELVVEHGGFVYDSDSYADDLPYWVRVHDTEHLVVPYTLDTN DMRFASPAGFSNGDEFFAHLRDAFDVLYAEGCDGSPKMLSVGLHCRLVGRPARIAALQRFLDHVQSHPRV WLARRIEIAEHWRAVHPARDREPRIQV*
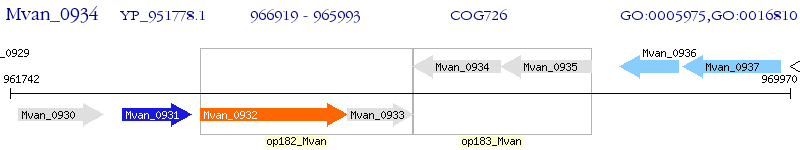
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0934 | - | - | 100% (308) | polysaccharide deacetylase |
| M. vanbaalenii PYR-1 | Mvan_3151 | - | 1e-69 | 45.61% (296) | polysaccharide deacetylase |
| M. vanbaalenii PYR-1 | Mvan_0047 | - | 9e-16 | 26.48% (287) | polysaccharide deacetylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0796 | - | 2e-14 | 27.24% (279) | polysaccharide deacetylase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2935c | - | 1e-142 | 79.33% (300) | polysaccharide deacetylase family protein |
| M. marinum M | MMAR_1211 | - | 1e-152 | 84.51% (297) | chitinase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5728 | - | 1e-164 | 90.30% (299) | polysaccharide deacetylase family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2681 | - | 1e-151 | 84.18% (297) | chitinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0934|M.vanbaalenii_PYR-1 MN-----------YPRDMVGYGRTPPDPQWPGGAAIAVQFVLNYEEGAEN
MSMEG_5728|M.smegmatis_MC2_155 MS-----------YPRDMVGYGRTPPDPQWPGGAAIAVQFVLNYEEGAEN
MMAR_1211|M.marinum_M MTG----------YPRDMTGYGRTPPDPQWPGGALIAVQFVLNYEEGAES
MUL_2681|M.ulcerans_Agy99 MTG----------YPRDMTGYGRTPPDPQWRGGALIAVQFVLNYEEGAES
MAB_2935c|M.abscessus_ATCC_199 MAHNEAHDEAPPATPRDMVGYGRTPPHPRWPGNARIAVQFVLNYEEGAEN
Mflv_0796|M.gilvum_PYR-GCK -----------------MDAFR-------WPDGKTAAAAFTFDVDA--ES
* .: * .. *. *.:: : *.
Mvan_0934|M.vanbaalenii_PYR-1 CVLDGDPASETFLSEITPAEAFPNRHMSMESLYEYGSRAGLWRVLRVFER
MSMEG_5728|M.smegmatis_MC2_155 CVLDGDPASETFLSEMTPAEAFPNRHMSMESLYEYGSRAGLWRVLRVFEQ
MMAR_1211|M.marinum_M SVLDGDSGSETFLSEIVPAQSFPNRHMSMESLYEYGSRAGVWRVLRVCEQ
MUL_2681|M.ulcerans_Agy99 SVLDGDSGSETFLSEIVPAQSFPNRHMSMESLYEYGSRAGVWRVLRVCEQ
MAB_2935c|M.abscessus_ATCC_199 NVLDGSAGSETFLSEMVPVEAFPNRHMSMESLYEYGSRAGLWRVLRVFER
Mflv_0796|M.gilvum_PYR-GCK AVLWGDES--------------VGARMSVMSHQAYGPLVGIPRILDLLEQ
** *. . . :**: * **. .*: *:* : *:
Mvan_0934|M.vanbaalenii_PYR-1 HDIPLTVFAVARALQRNPEAVAAFTELGHEIACHGLRWKSYQLTDRDTER
MSMEG_5728|M.smegmatis_MC2_155 RGIPLTIFAVARAMQRNPEAVAAFTELGHEIACHGLRWKSYQLTDRDTER
MMAR_1211|M.marinum_M SGVPLTIFAVAQAMQRNPEVAKVFRELGHEIACHGLRWKSYQEIDRDTER
MUL_2681|M.ulcerans_Agy99 SGVPLTIFAVAQAMQRNPEVAKGFRELGHEIACHGLRWKSYQEIDRDTER
MAB_2935c|M.abscessus_ATCC_199 RGLPLTIFAVATALQRNPEAVSAFGELGHEIACHGLRWQSYQLVPPDIER
Mflv_0796|M.gilvum_PYR-GCK HQITSTFFVPGHTADRYPEAVRSIVGAGHEIAHHGYLHEQPTALTLEEEI
:. *.*. . : :* **.. : ***** ** :. : *
Mvan_0934|M.vanbaalenii_PYR-1 VHLAEAVQILQDLTGSTPSGWYT-GRDSPHTRELVVEHGGFVYDSDSYAD
MSMEG_5728|M.smegmatis_MC2_155 EHMAEAVRILKELTGSAPLGWYT-GRDSPYTRELVVEHGGFVYDSDSYAD
MMAR_1211|M.marinum_M AHLSEAVRIHHELTGSAPRGWYG-GRDSPHTRELVVEHGGFLYDSDSYAD
MUL_2681|M.ulcerans_Agy99 AHLSEAVRIHHELTGSAPRGWYG-GRDSPHTRELVVEHGGFLYDSDSYAD
MAB_2935c|M.abscessus_ATCC_199 EHMAQAVQILRDLTGQAPLGWYT-GRDSPQTRQLVVEHGGFSYDSDSYAD
Mflv_0796|M.gilvum_PYR-GCK DALDRGLAALADVAGVRPAGYRAPMWDLSWRTPALLAERGFLYDSSLMDA
: ..: :::* * *: * . :: . ** ***.
Mvan_0934|M.vanbaalenii_PYR-1 DLPYWVRVHD----TEHLVVPYTLDTNDMRFASPAGFSNGDEFFAHLRDA
MSMEG_5728|M.smegmatis_MC2_155 DLPYWVQVDG----SDHLVVPYTLDTNDMRFASPAGFANGEEFFAHLRDA
MMAR_1211|M.marinum_M DLPYWVRVGA----SDHLVVPYTLDTNDMRFASPAGFSNGDEFFAHLRDA
MUL_2681|M.ulcerans_Agy99 DLPYWVRVGA----SDHLVVPYTLDTNDMRFASPAGFSNGDEFFAHLRDA
MAB_2935c|M.abscessus_ATCC_199 DLPYWVRVPAPGGVADHLVVPYTLDTNDMRFSSPAGFPSGEQFFTHLRDA
Mflv_0796|M.gilvum_PYR-GCK DHPYELAVTPG-AAQSLVEIPIQWALDDWEQYCFLPDISGSGLIESPRKA
* ** : * . : :* :* . . .*. :: *.*
Mvan_0934|M.vanbaalenii_PYR-1 FDVLYAEGCDGSPK--MLSVGLHCRLVGRPARIAALQRFLDHVQSHPRVW
MSMEG_5728|M.smegmatis_MC2_155 FDVLYAEGLAGAPK--MLSVGLHCRLVGRPARTAALQRFLDHVQSHDKVW
MMAR_1211|M.marinum_M FDVLYAEGRAGSPK--MLSVGLHCRLVGRPARTAALQRFLDHVRSHDRVW
MUL_2681|M.ulcerans_Agy99 FDVLYAEGRAGSPK--MLSVGLHCRLVGRPARTAALQRFLDHVRSHDRVW
MAB_2935c|M.abscessus_ATCC_199 FDCLYAEGEAGSPK--MLSVGLHCRLVGRPARTVALERFVDHVQAHDGVW
Mflv_0796|M.gilvum_PYR-GCK RELWQLEFDALRRVGGCWVLTNHPFLTGRPSRAAELGELMRYVLDHDDVW
: * : * *.***:* . * .:: :* * **
Mvan_0934|M.vanbaalenii_PYR-1 LARRIEIAEHWRAVHPARDREPRIQV--
MSMEG_5728|M.smegmatis_MC2_155 LARRIEIAEHWRTVHPA-----------
MMAR_1211|M.marinum_M LARRVEIAEHWREVHPAPTG--------
MUL_2681|M.ulcerans_Agy99 LARRVEIAEHWREVHPAPTG--------
MAB_2935c|M.abscessus_ATCC_199 ITKRIDIAEHWRRVHPAGTEPNESSSPA
Mflv_0796|M.gilvum_PYR-GCK VVSQA-----------------------
:. :