For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAETATLILLRHGESEWNALNLFTGWVDVDLTEKGRAEAARAGELLKAQDRLPDVLYTSLLRRAITTANI ALDKADRHWIPVHRDWRLNERHYGALQGLDKAETKAKYGEEQFMAWRRSYDTPPPPIEAGSEFSQDRDPR YADVVGGAPLTECLKDVVERFVPYFTEVIVPDLKAGKTVLIAAHGNSLRALVKYLDGMSDEDVVGLNIPT GIPLRYDLDADLKPTVPGGSYLDPEAAAAGAAAVAAQGAK
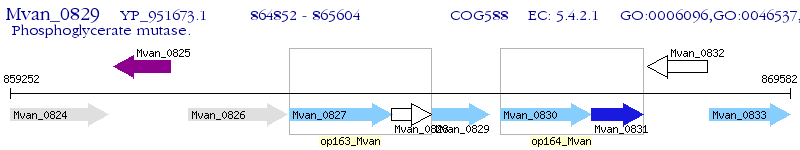
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0829 | - | - | 100% (250) | phosphoglycerate mutase 1 family protein |
| M. vanbaalenii PYR-1 | Mvan_3924 | - | 1e-14 | 32.99% (197) | phosphoglycerate mutase |
| M. vanbaalenii PYR-1 | Mvan_1788 | - | 1e-07 | 41.10% (73) | acid phosphatase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0499 | gpmA | 1e-116 | 80.24% (248) | phosphoglyceromutase |
| M. gilvum PYR-GCK | Mflv_0084 | - | 1e-135 | 92.80% (250) | phosphoglycerate mutase 1 family protein |
| M. tuberculosis H37Rv | Rv0489 | gpmA | 1e-116 | 80.24% (248) | phosphoglyceromutase |
| M. leprae Br4923 | MLBr_02441 | gpmA | 1e-113 | 81.78% (236) | phosphoglyceromutase |
| M. abscessus ATCC 19977 | MAB_4049c | - | 1e-119 | 83.33% (246) | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase |
| M. marinum M | MMAR_0814 | gpmA | 1e-118 | 83.13% (249) | phosphoglycerate mutase 1 Gpm1 |
| M. avium 104 | MAV_4662 | gpmA | 1e-120 | 83.87% (248) | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase |
| M. smegmatis MC2 155 | MSMEG_0935 | - | 1e-121 | 85.37% (246) | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase |
| M. thermoresistible (build 8) | TH_0879 | gpm | 1e-114 | 82.16% (241) | PROBABLE PHOSPHOGLYCERATE MUTASE 1 GPM1 |
| M. ulcerans Agy99 | MUL_4558 | gpmA | 1e-119 | 83.13% (249) | phosphoglycerate mutase 1 Gpm1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0499|M.bovis_AF2122/97 -------MANTGSLVLLRHGESDWNALNLFTGWVDVGLTDKGQAEAVRSG
Rv0489|M.tuberculosis_H37Rv -------MANTGSLVLLRHGESDWNALNLFTGWVDVGLTDKGQAEAVRSG
MMAR_0814|M.marinum_M -------MGDTGTLVLLRHGESDWNARNLFTGWVDVGLTEKGRAEAVRSG
MUL_4558|M.ulcerans_Agy99 -------MGDTGTLVLLRHGESDWNARNLFTGWVDVGLTEKGRAEAVRSG
MAV_4662|M.avium_104 -------MGETATLVLLRHGESEWNSLNLFTGWVDVGLTDKGRAEAVRSG
MLBr_02441|M.leprae_Br4923 -----MQQGNTATLILLRHGESDWNARNLFTGWVDVGLTDKGRAEAVRSG
Mvan_0829|M.vanbaalenii_PYR-1 -------MAETATLILLRHGESEWNALNLFTGWVDVDLTEKGRAEAARAG
Mflv_0084|M.gilvum_PYR-GCK -------MADTATLILLRHGESEWNALNLFTGWVDVDLTDKGRAEATRAG
MAB_4049c|M.abscessus_ATCC_199 MSGETSSKTNGATLILLRHGESQWNAKNLFTGWVDVDLTEKGRSEAQRGG
MSMEG_0935|M.smegmatis_MC2_155 ----------MATLILLRHGESQWNEKNLFTGWVDVDLTDKGRAEAVRGG
TH_0879|M.thermoresistible__bu MG-------NGATLILLRHGESDWNAKNLFTGWVDVDLTDKGRAEAVRAG
.:*:*******:** *********.**:**::** *.*
Mb0499|M.bovis_AF2122/97 ELIAEHDLLPDVLYTSLLRRAITTAHLALDSADRLWIPVRRSWRLNERHY
Rv0489|M.tuberculosis_H37Rv ELIAEHDLLPDVLYTSLLRRAITTAHLALDSADRLWIPVRRSWRLNERHY
MMAR_0814|M.marinum_M ELLAEQDLLPDVLYTSLLRRAITTAHLALDSADRLWIPVRRSWRLNERHY
MUL_4558|M.ulcerans_Agy99 ELLAEQDLLPDVLYTSLLRRAITTAHLALDSADRLWIPVRRSWRLNERHY
MAV_4662|M.avium_104 ELLAEQGLLPDVLYTSLLRRAITTAHLALDAADRLWIPVRRSWRLNERHY
MLBr_02441|M.leprae_Br4923 ELLAEHNLLPDVLYTSLLRRAITTAHLALDTADWLWIPVRRSWRLNERHY
Mvan_0829|M.vanbaalenii_PYR-1 ELLKAQDRLPDVLYTSLLRRAITTANIALDKADRHWIPVHRDWRLNERHY
Mflv_0084|M.gilvum_PYR-GCK ELIGEQDKLPDVLYTSLLRRAITTANIALDKADRHWIPVHRDWRLNERHY
MAB_4049c|M.abscessus_ATCC_199 ELLAQQGLLPDILFTSLLRRAINTAHLALDTADRLWIPVVRDWRLNERHY
MSMEG_0935|M.smegmatis_MC2_155 QLMAEQGVLPDVLYTSLLRRAITTAHLALDAADRLWIPVHRDWRLNERHY
TH_0879|M.thermoresistible__bu ELMAELDRLPDVVYTSLLRRAITTANLALDRADRHWIPVHRSWRLNERHY
:*: . ***:::********.**::*** ** **** *.********
Mb0499|M.bovis_AF2122/97 GALQGLDKAETKARYGEEQFMAWRRSYDTPPPPIERGSQFSQDADPRYAD
Rv0489|M.tuberculosis_H37Rv GALQGLDKAETKARYGEEQFMAWRRSYDTPPPPIERGSQFSQDADPRYAD
MMAR_0814|M.marinum_M GALQGLDKAETKARYGEEQFMAWRRSYDTPPPLIEKGSEFSQDADPRYAN
MUL_4558|M.ulcerans_Agy99 GALQGLDKAETKARYGEEQFMAWRRSYDTPPPLIEKGSEFSQDADPRYAN
MAV_4662|M.avium_104 GALQGLDKAETKARYGEEQFMAWRRSYDTPPPPIERGSTYSQDADPRYAD
MLBr_02441|M.leprae_Br4923 GALQGLDKAVTKARYGEERFMAWRRSYDTPPPPIEKGSEFSQDADPRYTD
Mvan_0829|M.vanbaalenii_PYR-1 GALQGLDKAETKAKYGEEQFMAWRRSYDTPPPPIEAGSEFSQDRDPRYAD
Mflv_0084|M.gilvum_PYR-GCK GALQGLNKAETKAKYGDEQFMAWRRSYDTPPPQIEAGSEFSQDRDPRYAD
MAB_4049c|M.abscessus_ATCC_199 GALQGLDKAETKAKYGEEQFMAWRRSYDTPPPPIERGSHYSQDQDPRYAD
MSMEG_0935|M.smegmatis_MC2_155 GALQGLDKAETKEKYGEEQFMAWRRSYDTPPPPIEKGSKYSQDADPRYAD
TH_0879|M.thermoresistible__bu GALQGLNKAETKEKYGEEQFMAWRRSYDTPPPPIERGSTYSQDGDPRYAD
******:** ** :**:*:************* ** ** :*** ****::
Mb0499|M.bovis_AF2122/97 IGGG-PLTECLADVVARFLPYFTDVIVGDLRVGKTVLIVAHGNSLRALVK
Rv0489|M.tuberculosis_H37Rv IGGG-PLTECLADVVARFLPYFTDVIVGDLRVGKTVLIVAHGNSLRALVK
MMAR_0814|M.marinum_M IDGG-PLTECLADVVARFLPYFTDVIVGDLRAGKTVLIVAHGNSLRALVK
MUL_4558|M.ulcerans_Agy99 IDGG-PLTECLADVVARFLPYFTDVIVGDLRAGKTVLIVAHGNSLRALVK
MAV_4662|M.avium_104 IGGG-PLTECLADVVVRFLPYFTDVIVPDLRSGKTVLIVAHGNSLRALVK
MLBr_02441|M.leprae_Br4923 IGGG-PLTECLADVVTRFLPYFTDVIVPDLRTGRTVLIVAHGNSLRALVK
Mvan_0829|M.vanbaalenii_PYR-1 VVGGAPLTECLKDVVERFVPYFTEVIVPDLKAGKTVLIAAHGNSLRALVK
Mflv_0084|M.gilvum_PYR-GCK IDGGAPLTECLKDVVERFVPYFTEVIVPDLKAGKTVLIAAHGNSLRALVK
MAB_4049c|M.abscessus_ATCC_199 IDGG-PLTECLADVVARFVPYYEEAIVPELRAGKTVLVAAHGNSLRALVK
MSMEG_0935|M.smegmatis_MC2_155 IDGG-PLTECLKDVVERFVPYYESAILPDLKAGKTVLIAAHGNSLRALVK
TH_0879|M.thermoresistible__bu IDGG-PLTECLADVVARFVPYYEQTIVPDLRAGKTVLIAAHGNSLRALVK
: ** ****** *** **:**: ..*: :*: *:***:.***********
Mb0499|M.bovis_AF2122/97 HLDQMSDDEIVGLNIPTGIPLRYDLDSAMRPLVRGGTYLDPEAAAAGAAA
Rv0489|M.tuberculosis_H37Rv HLDQMSDDEIVGLNIPTGIPLRYDLDSAMRPLVRGGTYLDPEAAAAGAAA
MMAR_0814|M.marinum_M HLDHMSDEDIVGLNIPTGIPLRYDLDERLQPVVPGGTYLDPEAAAAGAAA
MUL_4558|M.ulcerans_Agy99 HLDQMSDEDIVGLNIPTGIPLRYDLDERLQPVVPGGTYLDPEAAAAGAAA
MAV_4662|M.avium_104 HLDQMSDDDVVGLNIPTGIPLRYDLDARLRPLVPGGTYLDPEAAAAGAAA
MLBr_02441|M.leprae_Br4923 HLDEMSDDEVVGLNVPTGIPLRYDLDADLRPVVPGGTYLDPEAAAA----
Mvan_0829|M.vanbaalenii_PYR-1 YLDGMSDEDVVGLNIPTGIPLRYDLDADLKPTVPGGSYLDPEAAAAGAAA
Mflv_0084|M.gilvum_PYR-GCK YLDGMSDDDVVGLNIPTGIPLRYDLDESLKPLVAGGSYLDPDAAAAGAAA
MAB_4049c|M.abscessus_ATCC_199 YLDGISDEEIVGLNIPTGIPLRYDLDENLKPVTPGGVYLDPEAAAAGAAA
MSMEG_0935|M.smegmatis_MC2_155 YLDGMSDEEVVGLNIPTGIPLRYDLDENLKPVKIGGEYLDPEAAAAGAAA
TH_0879|M.thermoresistible__bu YLDGMSDEEVVGLNIPTGIPLRYDLDENLKPKVVGGQYLDXGPAWANSG-
:** :**:::****:*********** ::* ** *** .* *
Mb0499|M.bovis_AF2122/97 VAGQGRG--
Rv0489|M.tuberculosis_H37Rv VAGQGRG--
MMAR_0814|M.marinum_M VASQGAAKA
MUL_4558|M.ulcerans_Agy99 VASQGAAKA
MAV_4662|M.avium_104 VASQGGG--
MLBr_02441|M.leprae_Br4923 VISQARP--
Mvan_0829|M.vanbaalenii_PYR-1 VAAQGAK--
Mflv_0084|M.gilvum_PYR-GCK VAAQGAK--
MAB_4049c|M.abscessus_ATCC_199 VANQGAK--
MSMEG_0935|M.smegmatis_MC2_155 VAAQGAKK-
TH_0879|M.thermoresistible__bu ---------