For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALALTAITDRVHFAQTDLVNWTLVAEDGGVLLIDAGYPGHREEVLESLGRLGFGATDVRAILLTHAHIDH FGSAMWFAREHGTPVYCHADEVGHAKREYLEQASPLAVAAHAWQPRWLKWSLTLIRKGGLDRTGIPTTAA LSEDIAAGLPGAPVCVPTPGHTGGHCSYVVDGVLVSGDALITGHPVSRRRGPQLLPSVFNHDEVACLRSL EALGMLDTEVMLPGHGPVWRGPVRDAVAQATTAEP*
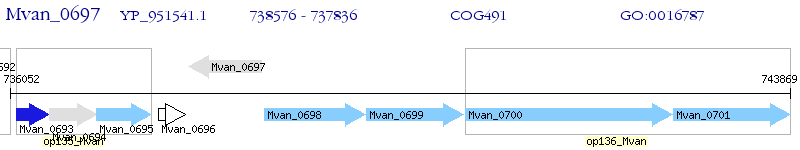
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0697 | - | - | 100% (246) | beta-lactamase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_2589 | - | 1e-06 | 31.28% (179) | beta-lactamase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1228 | - | 2e-05 | 27.23% (202) | beta-lactamase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0414c | - | 4e-89 | 63.29% (237) | beta lactamase like protein |
| M. gilvum PYR-GCK | Mflv_0212 | - | 1e-112 | 75.41% (244) | beta-lactamase domain-containing protein |
| M. tuberculosis H37Rv | Rv0406c | - | 4e-89 | 63.29% (237) | beta lactamase like protein |
| M. leprae Br4923 | MLBr_00270 | - | 2e-94 | 65.00% (240) | hypothetical protein MLBr_00270 |
| M. abscessus ATCC 19977 | MAB_4231 | - | 2e-73 | 52.77% (235) | Beta-lactamase-like protein |
| M. marinum M | MMAR_0708 | - | 5e-88 | 61.18% (237) | beta lactamase-like protein |
| M. avium 104 | MAV_4762 | - | 1e-94 | 64.98% (237) | metallo-beta-lactamase superfamily protein |
| M. smegmatis MC2 155 | MSMEG_0775 | - | 1e-104 | 68.70% (246) | metallo-beta-lactamase superfamily protein |
| M. thermoresistible (build 8) | TH_2303 | - | 2e-99 | 68.72% (243) | BETA LACTAMASE LIKE PROTEIN |
| M. ulcerans Agy99 | MUL_2815 | - | 4e-88 | 61.18% (237) | beta lactamase-like protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0697|M.vanbaalenii_PYR-1 -------------------MALALTAITDRVHFAQTDLVNWTLVAEDGGV
Mflv_0212|M.gilvum_PYR-GCK -------------------MAPVVTAVTDRVHFVRTELVNWTLVRDDDGV
MSMEG_0775|M.smegmatis_MC2_155 -------------------MAVALNKITDRVHFARTELVNWTLVTGDDGV
TH_2303|M.thermoresistible__bu ----------------MAAPAVRLEQITDSVHFANTDLVNWVLVTDGTGV
MLBr_00270|M.leprae_Br4923 ------------------MAAATLFQVTDSVHFARGDAVNWALVADDTGV
MAV_4762|M.avium_104 --------------------MAELVQVTDAVHLARGDAVNWTLVADDTGV
Mb0414c|M.bovis_AF2122/97 MVATRGTRLAALALAPRLAGMAELVQITDKVHLARGHAVNWVLVTDDTGV
Rv0406c|M.tuberculosis_H37Rv MVATRGTRLAALALAPRLAGMAELVQITDKVHLARGHAVNWVLVTDDTGV
MMAR_0708|M.marinum_M MPHQR----------------TELVQVTDKVHLAQGPAVNWVLVTDETGV
MUL_2815|M.ulcerans_Agy99 MPHQR----------------TELVQVTDKVHLAQGPAVNWVLVTDETGV
MAB_4231|M.abscessus_ATCC_1997 --------------------MVEITQVSDAVHVVNGEAVNWVIASDDSGV
: ::* **... ***.:. **
Mvan_0697|M.vanbaalenii_PYR-1 LLIDAGYPGHREEVLESLGRLGFGATDVRAILLTHAHIDHFGSAMWFARE
Mflv_0212|M.gilvum_PYR-GCK ILIDAGFPGHRDEVLTSLHGLGFGVGDVRAILLTHAHIDHFGTAIWFAAT
MSMEG_0775|M.smegmatis_MC2_155 LLIDAGFPGQREDVLDSVRQLGFGPDDIKAVLLTHAHVDHFGTAIWLAKT
TH_2303|M.thermoresistible__bu VLIDAGYPGQRGLVLHSLRQLGFGPDAVRAILLTHAHIDHLGSAIWFAKT
MLBr_00270|M.leprae_Br4923 MLIDTGYPGDRDDVVASLRQLGYEAGDVRAILLTHAHIDHMGSAIWFASK
MAV_4762|M.avium_104 MLVDAGYPGDREDVLGSLAELGYGPGDVRAIVLTHAHIDHLGTAIWLASE
Mb0414c|M.bovis_AF2122/97 LLIDAGYPGDRAEVLASLNKLGYTPGDVRAIVLTHAHIDHLGSAIWFARE
Rv0406c|M.tuberculosis_H37Rv LLIDAGYPGDRAEVLASLNKLGYTPGDVRAIVLTHAHIDHLGSAIWFARE
MMAR_0708|M.marinum_M MLIDAGYPGDRNNVLTSLRTLGYGPGDVRAILLTHAHIDHLGTAIWFAAQ
MUL_2815|M.ulcerans_Agy99 MLIDAGYPGDRNNVLTSLRTLGYGPGDVRAILLTHAHIDHLGTAIWFAAQ
MAB_4231|M.abscessus_ATCC_1997 TLFDSGYPGDRDDVLKSIAALGHKPQDVRAVVLTHGHIDHMGTAIWWAAE
*.*:*:**.* *: *: **. ::*::***.*:**:*:*:* *
Mvan_0697|M.vanbaalenii_PYR-1 HGTPVYCHADEVGHAKREYLEQASPLAVAAHAWQPRWLKWSLTLIRKGGL
Mflv_0212|M.gilvum_PYR-GCK HGTPVYCHADEVAHSRRDHLEQASPIDIAKHAWQPTWLKWSVSIIGKGAL
MSMEG_0775|M.smegmatis_MC2_155 YGTPVYCHADEVGHARREYLEQATPAGVARQAWQPRWLKWALAISRQGAF
TH_2303|M.thermoresistible__bu HGTPVFCHADEVGHAKRAYLEQVSPLDLAGQLWRPRWLTWSLTAVRTGAL
MLBr_00270|M.leprae_Br4923 HRTPVYCHADEVGHTKREYLEQVSLLNIAMRVWQPRWAIWSGHVLRSGGL
MAV_4762|M.avium_104 HGIPVYCHADEVGHAKREYLEQVSIVDVALRIWRPRWAKWTAHVVRSGGL
Mb0414c|M.bovis_AF2122/97 HSTPVYCHAEEVGHAKREYRENASVFDVALRSWRPRVAVWGIHLLRRGGL
Rv0406c|M.tuberculosis_H37Rv HSTPVYCHAEEVGHAKREYRENASVFDVALRSWRPRVAVWGIHLLRRGGL
MMAR_0708|M.marinum_M HGTPVYCDANEVGHAKREYHESASPLDVVLRSWRPRTALWGIHLVFRGGL
MUL_2815|M.ulcerans_Agy99 HGTPVYCDANEVGHAKREYHESASPLDVVLRSWRPRTALWGIHLVFRGGL
MAB_4231|M.abscessus_ATCC_1997 HGVPVYAHNAELGNVRRDYVDQAEPLKVVLNLWRPGWLPWVIHAMRLGAG
: **:.. *:.: :* : :.. :. . *:* * *.
Mvan_0697|M.vanbaalenii_PYR-1 DRTGIPTTAALSEDIAAGLPGAPVCVPTPGHTGGHCSYVVDG-VLVSGDA
Mflv_0212|M.gilvum_PYR-GCK TRAGIASTAPLTDHIAAALPGSPVCIPTPGHTGGHCSYVVDG-VLVSGDA
MSMEG_0775|M.smegmatis_MC2_155 DHSGIPSTQAFTPEIG--LPGNPTAIPTPGHTGGHCSYLVDG-VLVAGDA
TH_2303|M.thermoresistible__bu VREGIPGTRALTEDVAAQLPGQPVVIPSPGHTAGHCSYLIGE-VLVSGDA
MLBr_00270|M.leprae_Br4923 IRGGIPTAQPLTAETAAGLPGHPMAIFTPGHTGGHCSYVVDG-VLISGDA
MAV_4762|M.avium_104 IRDGIPTAQPLTAEVAAGLPGRPTPVFSPGHTNGHCSYLIDG-VLVSGDA
Mb0414c|M.bovis_AF2122/97 TGDGIPTAQPLTAEAAAGLPGQPMAIFTPGHTSGHCSYVVDG-VLASGDA
Rv0406c|M.tuberculosis_H37Rv TGDGIPTAQPLTAEAAAGLPGQPMAIFTPGHTSGHCSYVVDG-VLASGDA
MMAR_0708|M.marinum_M RRAGVPSTQALSAEVAAALPGHPIPIPTPGHTSGHCSYLVDG-VLASGDA
MUL_2815|M.ulcerans_Agy99 RRAGVPSTQALSAEVAAALPGHPIPIPTPGHTSGHCSYLVDG-VLASGDA
MAB_4231|M.abscessus_ATCC_1997 VRDGIPTAAPLGPELAD-LPGAPAPIPTPGHSGGHCSFLVAGHVLVVGDA
*:. : .: . . *** * : :***: ****::: ** ***
Mvan_0697|M.vanbaalenii_PYR-1 LITGHPVSRRRGPQLLPSVFNHDEVACLRSLEALGMLDTEVMLPGHGPVW
Mflv_0212|M.gilvum_PYR-GCK LITGHPVASRRGPQLLPSVFNHDEDACRRSLTTLGALDTEVILPGHGPLW
MSMEG_0775|M.smegmatis_MC2_155 LVTGHPTSSRRGPQLLHQVFNHDQQACLRSLGALGLLDTEVLLPGHGDVW
TH_2303|M.thermoresistible__bu LVTGHPVAPRRGPQLLPEVFNHDQDSCVRSVAALGLLDAEVLLPGHGPVW
MLBr_00270|M.leprae_Br4923 LITGHPLLRHRGPQLLTAVFSHSQKQCLRSLDALGLLETRILAPGHGELW
MAV_4762|M.avium_104 LITGHPLLRHRGPQLLPAIFSYSQRDCIRTLSALALLDTEVLLPGHGDVW
Mb0414c|M.bovis_AF2122/97 LITGHPMLRHRGPQLLPAVFSHSQQNSIRSLAALALLETNILAPGHGELW
Rv0406c|M.tuberculosis_H37Rv LITGHPMLRHRGPQLLPAVFSHSQQNSIRSLAALALLETNILAPGHGELW
MMAR_0708|M.marinum_M LITGHPMLRHRGPQLLPAVFSKVQERCIRSLSTLAQVEADTLAPGHGSLW
MUL_2815|M.ulcerans_Agy99 LITGHPMLRHRGPQLLPAVFSKVQERCIRSLSTLAQVEADTLAPGHGSLW
MAB_4231|M.abscessus_ATCC_1997 IITGHPVAKRSGPQLLPTPFTKDMEQARRSAEALADVDADVLAPGHGPAW
::**** : ***** *. . *: :*. ::: : **** *
Mvan_0697|M.vanbaalenii_PYR-1 RGPVRDAVAQATTAEP-------
Mflv_0212|M.gilvum_PYR-GCK RGPVRDAAAQAAQASVR------
MSMEG_0775|M.smegmatis_MC2_155 RGSIREMADQAASSAPSPAR---
TH_2303|M.thermoresistible__bu RGEISTAARLATPA---------
MLBr_00270|M.leprae_Br4923 HGPIREATEAALKQAGALTR---
MAV_4762|M.avium_104 RGPIKEATDAALALATGR-----
Mb0414c|M.bovis_AF2122/97 HGPIRKATDEALERAQKSNHVFR
Rv0406c|M.tuberculosis_H37Rv HGPIRKATDEALERAQKSNHVFR
MMAR_0708|M.marinum_M RGPIRLATEEARARAQR------
MUL_2815|M.ulcerans_Agy99 RGPIRLATEEARARAQR------
MAB_4231|M.abscessus_ATCC_1997 IGSLRDAVQIALNT---------
* : . *