For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VQDVDRVALEASIAVDVRALTAESDRIGHRFAGLHALTPNDFRALLHVMVAENAGTPLTAGELRNRMGTS GAAITYLVERMNASGHLRRESDPRDRRKVMLRVADHGMDVAREFFTPLSDHVHEGLAGLPDGDLAAAHRV FTALIDAMSTFRAALGADSGGSSGRLENR
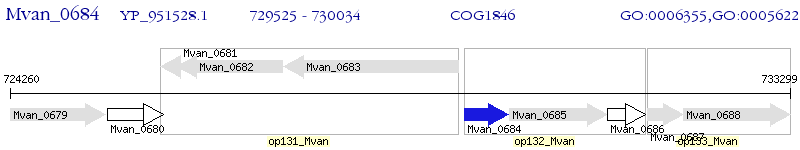
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0684 | - | - | 100% (169) | MarR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0537 | - | 5e-06 | 28.75% (80) | MarR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0148 | - | 2e-05 | 30.36% (112) | MarR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0223 | - | 3e-56 | 68.86% (167) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2648c | - | 2e-06 | 29.36% (109) | MarR family transcriptional regulator |
| M. marinum M | MMAR_1675 | - | 3e-46 | 57.69% (156) | transcriptional regulator |
| M. avium 104 | MAV_3893 | - | 3e-44 | 55.77% (156) | transcriptional regulator, MarR family protein |
| M. smegmatis MC2 155 | MSMEG_5566 | - | 4e-06 | 45.65% (46) | transcriptional regulator |
| M. thermoresistible (build 8) | TH_3314 | - | 3e-07 | 27.64% (123) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_0379 | - | 8e-40 | 49.38% (160) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0684|M.vanbaalenii_PYR-1 -----------MQDVDRVALEASIAVDVRALTAESDRIGHRFAGLHALTP
Mflv_0223|M.gilvum_PYR-GCK -----------MQNGDRAQVEALIAADVRALSAASDAIGQSFAALHGLTA
MMAR_1675|M.marinum_M --MLHIAMDADEQACSRQGLESAISADLRELSTESDQIGHLFAVSHSVRP
MAV_3893|M.avium_104 ----MTKGGAADRRLDRTELEKLMSADMRAITAQSDRIGRHFARQNNVSG
MUL_0379|M.ulcerans_Agy99 ----MTKRVAVNRQPSRAELERLLSADLRSITAHSDRVGRYYARHNDLSH
TH_3314|M.thermoresistible__bu -----------VTTSDRPQILAELNAEFRVQALRSVMLHSAVAARLGIAV
MSMEG_5566|M.smegmatis_MC2_155 -----------------MVAVQLVTSDLVGVALRS-------VDGLEVSL
MAB_2648c|M.abscessus_ATCC_199 MAVHESSITTRDWTLNRPELDTSPMEVIGALKRATGMLARLLEPLYATSA
. :
Mvan_0684|M.vanbaalenii_PYR-1 NDFRALLHVMVAENAGTPLTAGELRNRMGTS-GAAITYLVERMNASGHLR
Mflv_0223|M.gilvum_PYR-GCK SDFRALLHVTVAEDDGAPLTAGELRARMGTS-GAAITYLVERMIESGHVL
MMAR_1675|M.marinum_M NDFRALLHIMVAETGGRPLTSGQLGERMGLS-AAAITYLVDRLIESGHIR
MAV_3893|M.avium_104 TDFHALLHIMVAETTGTPLTPAQLRQRMDVS-PAAITYLVDRMIDAGHIR
MUL_0379|M.ulcerans_Agy99 GDFQALLHIMVAETAGTPVTPAQLSQRLDVS-AAAITYLADRMIHAGHIR
TH_3314|M.thermoresistible__bu TDFNCLNVLSMEG----PLTAGQLAERTDLTRGGAVTAMIDRLEAAGFVH
MSMEG_5566|M.smegmatis_MC2_155 PQFRLLRVLDEVGEAG----ATRCAEMLGIG-GSSITRLVDRLHASGHLV
MAB_2648c|M.abscessus_ATCC_199 IAEPEHDVLVVLRHRKGPVIARHVAEELGVS-QAWVSRMLRKLEERGYVR
: . . . . :: : :: *.:
Mvan_0684|M.vanbaalenii_PYR-1 RESDPRDRRKVMLRVADHGMDVAREFFTPLSDHVHEGLAGLPDGDLAAAH
Mflv_0223|M.gilvum_PYR-GCK RDTDPTDRRKVMLRVADHGMAVARDFFSPLSAHTHAALADLPDEDLLAAH
MMAR_1675|M.marinum_M RDTHPDDRRKVVLRYSEPGLATARSFFTPLGMHTRAALDGVADPDLVAAH
MAV_3893|M.avium_104 REPDPQDRRKTLLRYEKPGMALAHSFFTPLGAELRDALAHLSDRDLAAAH
MUL_0379|M.ulcerans_Agy99 READPDDRRKSHLRFEQSSLELAHRFFMPLGDHMREAMADLSDRDLRAAH
TH_3314|M.thermoresistible__bu RRRDDDDRRRVLVELDEAAAARIAPLFAGLGESLNDHLDSYRTDELRLLL
MSMEG_5566|M.smegmatis_MC2_155 RRPDPANRSAVVLVLTDEGRRLVRRVESRRRRELGKALDRLSPKERSQCI
MAB_2648c|M.abscessus_ATCC_199 REANANDRRAAIITMTDSGRSVVDELFPERLRIEAEALAGLGD-ERAAVV
* . :* : . . . : :
Mvan_0684|M.vanbaalenii_PYR-1 RVFTALIDAMSTFRAALGAD---SGGSSGRLENR---------
Mflv_0223|M.gilvum_PYR-GCK RVFAALTAAMVSFGAELG---------SGRLKTDD--------
MMAR_1675|M.marinum_M RVFTALIAAMRVFRTDLS---------SP--------------
MAV_3893|M.avium_104 RVFTAMIEAMTAFESRLAASTSKPPAAPGRKRATTAGRRGAPR
MUL_0379|M.ulcerans_Agy99 RVLTAMMYGMETFEKELYES---PAPTSGRPRKTPAQAESP--
TH_3314|M.thermoresistible__bu DAVRGINQRVARATDALREG-----------------------
MSMEG_5566|M.smegmatis_MC2_155 AALERLHEVLQAPEDPRIPY-----------------------
MAB_2648c|M.abscessus_ATCC_199 AGLERLVATLKAYEG----------------------------
. : :