For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLHIAMDADEQACSRQGLESAISADLRELSTESDQIGHLFAVSHSVRPNDFRALLHIMVAETGGRPLTSG QLGERMGLSAAAITYLVDRLIESGHIRRDTHPDDRRKVVLRYSEPGLATARSFFTPLGMHTRAALDGVAD PDLVAAHRVFTALIAAMRVFRTDLSSP
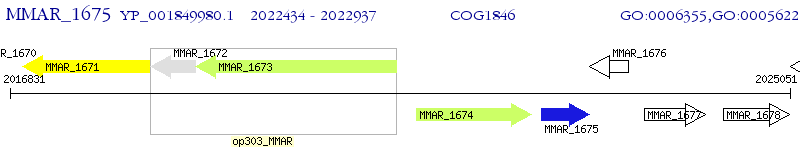
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1675 | - | - | 100% (167) | transcriptional regulator |
| M. marinum M | MMAR_4811 | - | 3e-39 | 51.66% (151) | MarR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0223 | - | 2e-44 | 58.55% (152) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0208 | - | 4e-05 | 32.43% (74) | MarR family transcriptional regulator |
| M. avium 104 | MAV_3893 | - | 9e-45 | 55.97% (159) | transcriptional regulator, MarR family protein |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3314 | - | 2e-08 | 29.23% (130) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_0379 | - | 2e-39 | 51.66% (151) | MarR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0684 | - | 3e-46 | 57.69% (156) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0223|M.gilvum_PYR-GCK ---------MQNGDRAQVEALIAADVR-------ALSAASDAIGQSFAAL
Mvan_0684|M.vanbaalenii_PYR-1 ---------MQDVDRVALEASIAVDVR-------ALTAESDRIGHRFAGL
MMAR_1675|M.marinum_M MLHIAMDADEQACSRQGLESAISADLR-------ELSTESDQIGHLFAVS
MAV_3893|M.avium_104 --MTKGGAADRRLDRTELEKLMSADMR-------AITAQSDRIGRHFARQ
MUL_0379|M.ulcerans_Agy99 --MTKRVAVNRQPSRAELERLLSADLR-------SITAHSDRVGRYYARH
TH_3314|M.thermoresistible__bu ---------VTTSDRPQILAELNAEFR-------VQALRSVMLHSAVAAR
MAB_0208|M.abscessus_ATCC_1997 ---------MSTRSESEVPGLDLAEFRSWQNFWVATNRLNFLLNRKMVAS
.. : .:.* . : .
Mflv_0223|M.gilvum_PYR-GCK HGLTASDFRALLHVTVAEDDGAPLTAGELRARMGTS-GAAITYLVERMIE
Mvan_0684|M.vanbaalenii_PYR-1 HALTPNDFRALLHVMVAENAGTPLTAGELRNRMGTS-GAAITYLVERMNA
MMAR_1675|M.marinum_M HSVRPNDFRALLHIMVAETGGRPLTSGQLGERMGLS-AAAITYLVDRLIE
MAV_3893|M.avium_104 NNVSGTDFHALLHIMVAETTGTPLTPAQLRQRMDVS-PAAITYLVDRMID
MUL_0379|M.ulcerans_Agy99 NDLSHGDFQALLHIMVAETAGTPVTPAQLSQRLDVS-AAAITYLADRMIH
TH_3314|M.thermoresistible__bu LGIAVTDFNCLNVLSMEG----PLTAGQLAERTDLTRGGAVTAMIDRLEA
MAB_0208|M.abscessus_ATCC_1997 HSISLTDFHVLQQLLRSANG--SCRMGDIANSLLAS-RSRITHQVRRLEE
: **. * : . .:: : . :* *:
Mflv_0223|M.gilvum_PYR-GCK SGHVLRDTDPTDRRKVMLRVADHGMAVARDFFSPLSAHTHAA-LADLPDE
Mvan_0684|M.vanbaalenii_PYR-1 SGHLRRESDPRDRRKVMLRVADHGMDVAREFFTPLSDHVHEG-LAGLPDG
MMAR_1675|M.marinum_M SGHIRRDTHPDDRRKVVLRYSEPGLATARSFFTPLGMHTRAA-LDGVADP
MAV_3893|M.avium_104 AGHIRREPDPQDRRKTLLRYEKPGMALAHSFFTPLGAELRDA-LAHLSDR
MUL_0379|M.ulcerans_Agy99 AGHIRREADPDDRRKSHLRFEQSSLELAHRFFMPLGDHMREA-MADLSDR
TH_3314|M.thermoresistible__bu AGFVHRRRDDDDRRRVLVELDEAAAARIAPLFAGLGESLNDH-LDSYRTD
MAB_0208|M.abscessus_ATCC_1997 QGLVKRGSMPQDRRAVLAALTEQGRTLGEAAMRTYSECVREHYLMRLTRP
* : * *** . . : . . :
Mflv_0223|M.gilvum_PYR-GCK DLLAAHRVFAALTAAMVSFGAELG---------SGRLKTDD--------
Mvan_0684|M.vanbaalenii_PYR-1 DLAAAHRVFTALIDAMSTFRAALGAD---SGGSSGRLENR---------
MMAR_1675|M.marinum_M DLVAAHRVFTALIAAMRVFRTDLS---------SP--------------
MAV_3893|M.avium_104 DLAAAHRVFTAMIEAMTAFESRLAASTSKPPAAPGRKRATTAGRRGAPR
MUL_0379|M.ulcerans_Agy99 DLRAAHRVLTAMMYGMETFEKELYES---PAPTSGRPRKTPAQAESP--
TH_3314|M.thermoresistible__bu ELRLLLDAVRGINQRVARATDALREG-----------------------
MAB_0208|M.abscessus_ATCC_1997 QQAAIAESFRRVGEGAEEALHDGDGR-------GGE-------------
: . :