For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTKGGAADRRLDRTELEKLMSADMRAITAQSDRIGRHFARQNNVSGTDFHALLHIMVAETTGTPLTPAQL RQRMDVSPAAITYLVDRMIDAGHIRREPDPQDRRKTLLRYEKPGMALAHSFFTPLGAELRDALAHLSDRD LAAAHRVFTAMIEAMTAFESRLAASTSKPPAAPGRKRATTAGRRGAPR
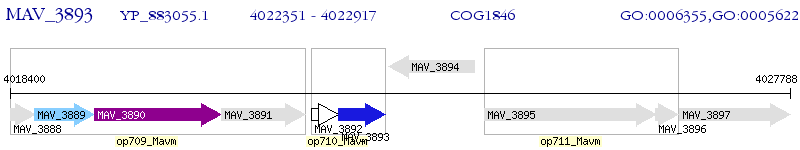
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3893 | - | - | 100% (188) | transcriptional regulator, MarR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0223 | - | 2e-41 | 52.23% (157) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1021 | - | 1e-05 | 30.00% (90) | MarR family transcriptional regulator |
| M. marinum M | MMAR_4811 | - | 3e-58 | 62.98% (181) | MarR family transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6913 | - | 2e-05 | 29.17% (96) | putative transcriptional regulatory protein |
| M. thermoresistible (build 8) | TH_3314 | - | 2e-09 | 26.92% (156) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_0379 | - | 2e-57 | 62.43% (181) | MarR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0684 | - | 4e-44 | 55.77% (156) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4811|M.marinum_M MTKRVAVNRQPSRAELERLLSADLRAITAHSDRVGRYYA-RHNDLSHGDF
MUL_0379|M.ulcerans_Agy99 MTKRVAVNRQPSRAELERLLSADLRSITAHSDRVGRYYA-RHNDLSHGDF
MAV_3893|M.avium_104 MTKGGAADRRLDRTELEKLMSADMRAITAQSDRIGRHFA-RQNNVSGTDF
Mflv_0223|M.gilvum_PYR-GCK MQN-------GDRAQVEALIAADVRALSAASDAIGQSFA-ALHGLTASDF
Mvan_0684|M.vanbaalenii_PYR-1 MQD-------VDRVALEASIAVDVRALTAESDRIGHRFA-GLHALTPNDF
TH_3314|M.thermoresistible__bu VTT-------SDRPQILAELNAEFRVQALRSVMLHSAVA-ARLGIAVTDF
MSMEG_6913|M.smegmatis_MC2_155 ---------MAETEPQVTELAGELQRVLSKVFSVLRRGD-THRGTAGDLT
MAB_1021|M.abscessus_ATCC_1997 --------MTVDDVDLDESLDALADSLLVASRLLLAISARSIAQVDETIT
. : :
MMAR_4811|M.marinum_M QALLHIMVAETAGTPVTPAQLSQRLDVS-AAAITYLADRMIHAGHIRREA
MUL_0379|M.ulcerans_Agy99 QALLHIMVAETAGTPVTPAQLSQRLDVS-AAAITYLADRMIHAGHIRREA
MAV_3893|M.avium_104 HALLHIMVAETTGTPLTPAQLRQRMDVS-PAAITYLVDRMIDAGHIRREP
Mflv_0223|M.gilvum_PYR-GCK RALLHVTVAEDDGAPLTAGELRARMGTS-GAAITYLVERMIESGHVLRDT
Mvan_0684|M.vanbaalenii_PYR-1 RALLHVMVAENAGTPLTAGELRNRMGTS-GAAITYLVERMNASGHLRRES
TH_3314|M.thermoresistible__bu NCLNVLSMEG----PLTAGQLAERTDLTRGGAVTAMIDRLEAAGFVHRRR
MSMEG_6913|M.smegmatis_MC2_155 LAQLSILLTLLEGGPIRMTELAARERVR-TPTTTVAIRRLEKLGLVKRSR
MAB_1021|M.abscessus_ATCC_1997 FPQFRTLVILFNEGPINLATLAQSLGVQ-PSTTGRMVDRLVSAGLIDRRP
: *: * : *: * : *
MMAR_4811|M.marinum_M DPDDRRKSHLRFEQSSLELAHKFFMPLGDHMREAMADLSDRDLRAAHRVL
MUL_0379|M.ulcerans_Agy99 DPDDRRKSHLRFEQSSLELAHRFFMPLGDHMREAMADLSDRDLRAAHRVL
MAV_3893|M.avium_104 DPQDRRKTLLRYEKPGMALAHSFFTPLGAELRDALAHLSDRDLAAAHRVF
Mflv_0223|M.gilvum_PYR-GCK DPTDRRKVMLRVADHGMAVARDFFSPLSAHTHAALADLPDEDLLAAHRVF
Mvan_0684|M.vanbaalenii_PYR-1 DPRDRRKVMLRVADHGMDVAREFFTPLSDHVHEGLAGLPDGDLAAAHRVF
TH_3314|M.thermoresistible__bu DDDDRRRVLVELDEAAAARIAPLFAGLGESLNDHLDSYRTDELRLLLDAV
MSMEG_6913|M.smegmatis_MC2_155 DPSDLRAVLVDVTPQGLVQHRESLAARRADLAERLSKLSDEDLATLARAL
MAB_1021|M.abscessus_ATCC_1997 HPRSRRELVAELTAHGEGVVREVMAQRRAEIAEIAARMPARERRGLVRAL
. . * . : : ..
MMAR_4811|M.marinum_M TAMMDGMETFEKELYES---PAPTSGRPRKTPAQAESP--
MUL_0379|M.ulcerans_Agy99 TAMMYGMETFEKELYES---PAPTSGRPRKTPAQAESP--
MAV_3893|M.avium_104 TAMIEAMTAFESRLAASTSKPPAAPGRKRATTAGRRGAPR
Mflv_0223|M.gilvum_PYR-GCK AALTAAMVSFGAELG---------SGRLKTDD--------
Mvan_0684|M.vanbaalenii_PYR-1 TALIDAMSTFRAALGAD---SGGSSGRLENR---------
TH_3314|M.thermoresistible__bu RGINQRVARATDALREG-----------------------
MSMEG_6913|M.smegmatis_MC2_155 GPLERLAGQDEDKKNER------ETPKPAEQTS-------
MAB_1021|M.abscessus_ATCC_1997 DAFTAAGGESASPVDV------------------------
: